[English] 日本語
 Yorodumi
Yorodumi- EMDB-63589: Imidazole glycerol phosphate dehydratase from Mycobacterium tuber... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Imidazole glycerol phosphate dehydratase from Mycobacterium tuberculosis, in complex with aminotriazole | |||||||||||||||
 Map data Map data | Combined, sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | histidine pathway / dehydratase / Mycobacterium tuberculosis / LYASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationimidazoleglycerol-phosphate dehydratase / imidazoleglycerol-phosphate dehydratase activity / L-histidine biosynthetic process / metal ion binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Raina R / Kar D / Singla M / Tiwari S / Kumari S / Aneja S / Kumar V / Banerjee S / Goyal S / Pal RK ...Raina R / Kar D / Singla M / Tiwari S / Kumari S / Aneja S / Kumar V / Banerjee S / Goyal S / Pal RK / Vinothkumar KR / Biswal BK | |||||||||||||||
| Funding support |  India, 4 items India, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Acta Crystallogr F Struct Biol Commun / Year: 2025 Journal: Acta Crystallogr F Struct Biol Commun / Year: 2025Title: Cryo-EM structures of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in the apo state and in the presence of small molecules. Authors: Rahul Raina / Deepsikha Kar / Mohini Singla / Satish Tiwari / Swati Kumari / Sonanjali Aneja / Varun Kumar / Soumya Banerjee / Shivika Goyal / Ravi Kant Pal / Kutti R Vinothkumar / Bichitra Biswal /  Abstract: Unlike humans, Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, has a de novo histidine-biosynthesis pathway. The enzyme imidazole glycerol phosphate dehydratase (IGPD), ...Unlike humans, Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, has a de novo histidine-biosynthesis pathway. The enzyme imidazole glycerol phosphate dehydratase (IGPD), which catalyses the conversion of imidazole glycerol phosphate to imidazole acetol phosphate, has been studied extensively from various organisms and has become a major target for the development of antibacterial, antiweed and antifungal small molecules. In our previous studies, we have shown that in crystals IGPD forms a 24-mer oligomeric state in which the monomers are arranged in 432 symmetry. In order to gain insights into the oligomeric state of Mtb IGPD in solution, we determined cryogenic sample electron microscopy (cryo-EM) structures of apo IGPD at 2.2 and 3.1 Å resolution. In addition, we also determined the cryo-EM structure of IGPD in the presence of 3-amino-1,2,4-triazole (ATZ) to 2.8 Å resolution. The results of this work, which corroborate those from the crystallographic studies, indicate that IGPD forms a homo-oligomeric structure in solution comprising of 24 subunits. ATZ binds in the active-site pocket of the enzyme, which is located at the interface of three monomers and tethers 24 ATZ molecules. The results of this study suggest that cryo-EM, in addition to being a rapidly evolving and complementary imaging technology for elucidating 3D structures of biological macromolecules, can be useful in pinpointing the mode of binding small molecules of low mass (here ∼85 Da) and mapping protein-ligand interactions, which could assist in the design of accurate (high-potency) inhibitors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63589.map.gz emd_63589.map.gz | 55.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63589-v30.xml emd-63589-v30.xml emd-63589.xml emd-63589.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_63589_fsc.xml emd_63589_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_63589.png emd_63589.png | 98 KB | ||
| Filedesc metadata |  emd-63589.cif.gz emd-63589.cif.gz | 6.8 KB | ||
| Others |  emd_63589_half_map_1.map.gz emd_63589_half_map_1.map.gz emd_63589_half_map_2.map.gz emd_63589_half_map_2.map.gz | 42.1 MB 41.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63589 http://ftp.pdbj.org/pub/emdb/structures/EMD-63589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63589 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63589 | HTTPS FTP |
-Related structure data
| Related structure data |  9m2qMC  9m2pC  9m2rC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_63589.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63589.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined, sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: One of the half maps
| File | emd_63589_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps
| File | emd_63589_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium tuberculosis HisB with 3TR
| Entire | Name: Mycobacterium tuberculosis HisB with 3TR |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium tuberculosis HisB with 3TR
| Supramolecule | Name: Mycobacterium tuberculosis HisB with 3TR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: An oligomer of 24 subunits |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 550 KDa |
-Macromolecule #1: Imidazoleglycerol-phosphate dehydratase
| Macromolecule | Name: Imidazoleglycerol-phosphate dehydratase / type: protein_or_peptide / ID: 1 Details: The enzyme was expressed with a Histag at the N-terminus but not observed in the experiment. The C-terminal residues are also not observed in the experiment. Uniparc ID is UPI000012C861. Number of copies: 24 / Enantiomer: LEVO / EC number: imidazoleglycerol-phosphate dehydratase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.633516 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MHHHHHHTTT QTAKASRRAR IERRTRESDI VIELDLDGTG QVAVDTGVPF YDHMLTALGS HASFDLTVRA TGDVEIEAHH TIEDTAIAL GTALGQALGD KRGIRRFGDA FIPMDETLAH AAVDLSGRPY CVHTGEPDHL QHTTIAGSSV PYHTVINRHV F ESLAANAR ...String: MHHHHHHTTT QTAKASRRAR IERRTRESDI VIELDLDGTG QVAVDTGVPF YDHMLTALGS HASFDLTVRA TGDVEIEAHH TIEDTAIAL GTALGQALGD KRGIRRFGDA FIPMDETLAH AAVDLSGRPY CVHTGEPDHL QHTTIAGSSV PYHTVINRHV F ESLAANAR IALHVRVLYG RDPHHITEAQ YKAVARALRQ AVEPDPRVSG VPSTKGAL UniProtKB: Imidazoleglycerol-phosphate dehydratase |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 48 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #3: 3-AMINO-1,2,4-TRIAZOLE
| Macromolecule | Name: 3-AMINO-1,2,4-TRIAZOLE / type: ligand / ID: 3 / Number of copies: 24 / Formula: 3TR |
|---|---|
| Molecular weight | Theoretical: 84.08 Da |
| Chemical component information | 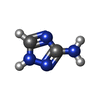 ChemComp-3TR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Enzyme was incubated with 10 fold excess of 3TR and this mixture was directly used for freezing |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Average exposure time: 60.0 sec. / Average electron dose: 27.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 130841 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 35.0 µm / Nominal defocus min: 12.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 10-199 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 133.9 |
| Output model |  PDB-9m2q: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)