+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PGE2-EP1-Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / EP1 / PGE2 / Gq / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprostaglandin E receptor activity / Prostanoid ligand receptors / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation / entrainment of circadian clock / regulation of canonical Wnt signaling pathway ...prostaglandin E receptor activity / Prostanoid ligand receptors / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation / entrainment of circadian clock / regulation of canonical Wnt signaling pathway / glutamate receptor signaling pathway / phototransduction, visible light / PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / photoreceptor outer segment / D1 dopamine receptor binding / intracellular transport / neuropeptide signaling pathway / postsynaptic cytosol / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase inhibitor activity / adenylate cyclase-activating adrenergic receptor signaling pathway / positive regulation of protein localization to cell cortex / enzyme regulator activity / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / cellular response to forskolin / GTPase activator activity / regulation of mitotic spindle organization / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / adenylate cyclase activator activity / trans-Golgi network membrane / Regulation of insulin secretion / negative regulation of inflammatory response to antigenic stimulus / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / bone development / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / cognition / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / blood coagulation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / sensory perception of smell / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Meng X / Xu Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structural insights into the activation of the human prostaglandin E receptor EP1 subtype by prostaglandin E. Authors: Xue Meng / Yang Li / Jiuyin Xu / Kai Wu / Wen Hu / Canrong Wu / H Eric Xu / Youwei Xu /  Abstract: Prostaglandin E (PGE) mediates diverse physiological processes through four G protein-coupled receptor subtypes (EP1-EP4). While structures of EP2, EP3, and EP4 have been determined, the structural ...Prostaglandin E (PGE) mediates diverse physiological processes through four G protein-coupled receptor subtypes (EP1-EP4). While structures of EP2, EP3, and EP4 have been determined, the structural basis for PGE recognition and activation of the EP1 receptor subtype has remained elusive due to its inherent instability. Here, we present the cryoelectron microscopy structure of the human EP1 receptor in complex with PGE and heterotrimeric Gq protein at 2.55 Å resolution, completing the structural characterization of the EP receptor family. Our structure reveals a unique binding mode of PGE within EP1, involving key interactions with residues in the orthosteric pocket. Notably, we observe a less pronounced outward displacement of transmembrane helix 6 compared to other EP receptor subtypes, suggesting a distinct activation mechanism for EP1. Through extensive mutational analyses, we identify critical residues involved in PGE recognition, EP1 activation, and Gq protein coupling. By overcoming the challenges associated with the instability of EP1, our findings provide valuable insights into the subtype-specific activation mechanisms of EP receptors and lay the foundation for the development of more selective EP1-targeted therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63571.map.gz emd_63571.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63571-v30.xml emd-63571-v30.xml emd-63571.xml emd-63571.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63571.png emd_63571.png | 37.8 KB | ||
| Filedesc metadata |  emd-63571.cif.gz emd-63571.cif.gz | 6.8 KB | ||
| Others |  emd_63571_half_map_1.map.gz emd_63571_half_map_1.map.gz emd_63571_half_map_2.map.gz emd_63571_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63571 http://ftp.pdbj.org/pub/emdb/structures/EMD-63571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63571 | HTTPS FTP |
-Related structure data
| Related structure data |  9m1hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_63571.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63571.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_63571_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63571_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PGE2-EP1-Gq complex
| Entire | Name: PGE2-EP1-Gq complex |
|---|---|
| Components |
|
-Supramolecule #1: PGE2-EP1-Gq complex
| Supramolecule | Name: PGE2-EP1-Gq complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Prostaglandin E2 receptor EP1 subtype
| Macromolecule | Name: Prostaglandin E2 receptor EP1 subtype / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.84723 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSPCGPLNLS LAGEATTCAA PWVPNTSAVP PSGASPALPI FSMTLGAVSN LLALALLAQA AGRLRRRRSA ATFLLFVASL LATDLAGHV IPGALVLRLY TAGRAPAGGA CHFLGGCMVF FGLCPLLLGC GMAVERCVGV TRPLLHAARV SVARARLALA A VAAVALAV ...String: MSPCGPLNLS LAGEATTCAA PWVPNTSAVP PSGASPALPI FSMTLGAVSN LLALALLAQA AGRLRRRRSA ATFLLFVASL LATDLAGHV IPGALVLRLY TAGRAPAGGA CHFLGGCMVF FGLCPLLLGC GMAVERCVGV TRPLLHAARV SVARARLALA A VAAVALAV ALLPLARVGR YELQYPGTWC FIGLGPPGGW RQALLAGLFA SLGLVALLAA LVCNTLSGLA LLRARWRRRS RR PPPASGP DSRRRWGAHG PRSASASSAS SIASASTFFG GSRSSGSARR ARAHDVEMVG QLVGIMVVSC ICWSPMLVLV ALA VGGWSS TSLQRPLFLA VRLASWNQIL DPWVYILLRQ AVLRQLLRLL PPRAGAKGGP AGLGLTPSAW EASSLRSSRH SGLS HF UniProtKB: Prostaglandin E2 receptor EP1 subtype |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine n...
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(q) subunit alpha type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.724383 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF KSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCSVDT ENARRIFNDC KDIILQMNLR EYNLV UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(i) subunit alpha-1, ...UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(i) subunit alpha-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(q) subunit alpha |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-...
| Macromolecule | Name: (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid type: ligand / ID: 5 / Number of copies: 1 / Formula: P2E |
|---|---|
| Molecular weight | Theoretical: 352.465 Da |
| Chemical component information | 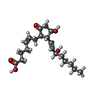 ChemComp-P2E: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)