[English] 日本語
 Yorodumi
Yorodumi- EMDB-63398: Cryo-EM structure of Arabidopsis thaliana RGA in complex with GID1A -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis thaliana RGA in complex with GID1A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Gibberellin / DELLA motif / GRAS domain / Plant growth / HORMONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationfruit morphogenesis / gibberellin mediated signaling pathway / positive regulation of gibberellic acid mediated signaling pathway / floral organ morphogenesis / negative regulation of trichome patterning / negative regulation of developmental vegetative growth / negative regulation of leaf development / regulation of seed dormancy process / gibberellin binding / negative regulation of gibberellic acid mediated signaling pathway ...fruit morphogenesis / gibberellin mediated signaling pathway / positive regulation of gibberellic acid mediated signaling pathway / floral organ morphogenesis / negative regulation of trichome patterning / negative regulation of developmental vegetative growth / negative regulation of leaf development / regulation of seed dormancy process / gibberellin binding / negative regulation of gibberellic acid mediated signaling pathway / positive regulation of fertilization / meiotic cytokinesis / response to gibberellin / gibberellic acid mediated signaling pathway / regulation of seed germination / response to far red light / Hydrolases / regulation of protein catabolic process / response to cold / promoter-specific chromatin binding / cellular response to hypoxia / transcription cis-regulatory region binding / hydrolase activity / DNA-binding transcription factor activity / positive regulation of gene expression / regulation of DNA-templated transcription / positive regulation of DNA-templated transcription / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Islam S / Park K / Kwon E / Kim DY | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Plant / Year: 2025 Journal: Mol Plant / Year: 2025Title: Structural insights into gibberellin-mediated DELLA protein degradation. Authors: Soyaab Islam / Kunwoong Park / Jing Xia / Eunju Kwon / Dong Young Kim /  Abstract: Gibberellin promotes plant growth by downregulating DELLA proteins, which act as growth repressors. In the presence of gibberellin, the gibberellin receptor GID1 binds DELLA proteins, triggering ...Gibberellin promotes plant growth by downregulating DELLA proteins, which act as growth repressors. In the presence of gibberellin, the gibberellin receptor GID1 binds DELLA proteins, triggering their degradation through polyubiquitination by the SCF ubiquitin E3 ligase. Despite extensive studies, the molecular mechanisms by which DELLA proteins assemble with SCF to regulate plant growth remain poorly understood. Here, we present two cryo-electron microscopy structures of the Arabidopsis thaliana DELLA protein RGA in complex with GID1A and GID1A-SLY1-ASK2, respectively. Structural analyses revealed that RGA interacts with GID1A and SLY1 through nonoverlapping binding surfaces, stabilizing the proteins. This suggests that the SCF-RGA-GID1A complex assembles through a stepwise stabilization process induced by gibberellin. Furthermore, structural comparison with GRAS proteins indicates that RGA does not interact with IDD-family transcription factors when bound to SLY1, suggesting that DELLA protein binding to GID1/SLY1 and to transcription factors is mutually exclusive. These findings provide new insights into the gibberellin-mediated regulation of transcription factor activity by DELLA proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_63398.map.gz emd_63398.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-63398-v30.xml emd-63398-v30.xml emd-63398.xml emd-63398.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_63398.png emd_63398.png | 152.6 KB | ||
| Masks |  emd_63398_msk_1.map emd_63398_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-63398.cif.gz emd-63398.cif.gz | 6.7 KB | ||
| Others |  emd_63398_half_map_1.map.gz emd_63398_half_map_1.map.gz emd_63398_half_map_2.map.gz emd_63398_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-63398 http://ftp.pdbj.org/pub/emdb/structures/EMD-63398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63398 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-63398 | HTTPS FTP |
-Related structure data
| Related structure data |  9lumMC  9lunC  9luoC  9lupC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_63398.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_63398.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7451 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_63398_msk_1.map emd_63398_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_63398_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_63398_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterodimer of RGA and GID1A/GA3
| Entire | Name: Heterodimer of RGA and GID1A/GA3 |
|---|---|
| Components |
|
-Supramolecule #1: Heterodimer of RGA and GID1A/GA3
| Supramolecule | Name: Heterodimer of RGA and GID1A/GA3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Heterodimer of the DELLA protein RGA and gibberellin receptor GID1A in complex with gibberellin A3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103 KDa |
-Macromolecule #1: DELLA protein RGA
| Macromolecule | Name: DELLA protein RGA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.113125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSKRDHHQFQ GRLSNHGTSS SSSSISKDKM MMVKKEEDGG GNMDDELLAV LGYKVRSSEM AEVALKLEQL ETMMSNVQED GLSHLATDT VHYNPSELYS WLDNMLSELN PPPLPASSNG LDPVLPSPEI CGFPASDYDL KVIPGNAIYQ FPAIDSSSSS N NQNKRLKS ...String: GSKRDHHQFQ GRLSNHGTSS SSSSISKDKM MMVKKEEDGG GNMDDELLAV LGYKVRSSEM AEVALKLEQL ETMMSNVQED GLSHLATDT VHYNPSELYS WLDNMLSELN PPPLPASSNG LDPVLPSPEI CGFPASDYDL KVIPGNAIYQ FPAIDSSSSS N NQNKRLKS CSSPDSMVTS TSTGTQIGGV IGTTVTTTTT TTTAAGESTR SVILVDSQEN GVRLVHALMA CAEAIQQNNL TL AEALVKQ IGCLAVSQAG AMRKVATYFA EALARRIYRL SPPQNQIDHC LSDTLQMHFY ETCPYLKFAH FTANQAILEA FEG KKRVHV IDFSMNQGLQ WPALMQALAL REGGPPTFRL TGIGPPAPDN SDHLHEVGCK LAQLAEAIHV EFEYRGFVAN SLAD LDASM LELRPSDTEA VAVNSVFELH KLLGRPGGIE KVLGVVKQIK PVIFTVVEQE SNHNGPVFLD RFTESLHYYS TLFDS LEGV PNSQDKVMSE VYLGKQICNL VACEGPDRVE RHETLSQWGN RFGSSGLAPA HLGSNAFKQA SMLLSVFNSG QGYRVE ESN GCLMLGWHTR PLITTSAWKL STAAY UniProtKB: DELLA protein RGA |
-Macromolecule #2: Gibberellin receptor GID1A
| Macromolecule | Name: Gibberellin receptor GID1A / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.808922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMAASDEVN LIESRTVVPL NTWVLISNFK VAYNILRRPD GTFNRHLAEY LDRKVTANAN PVDGVFSFDV LIDRRINLLS RVYRPAYAD QEQPPSILDL EKPVDGDIVP VILFFHGGSF AHSSANSAIY DTLCRRLVGL CKCVVVSVNY RRAPENPYPC A YDDGWIAL ...String: GSMAASDEVN LIESRTVVPL NTWVLISNFK VAYNILRRPD GTFNRHLAEY LDRKVTANAN PVDGVFSFDV LIDRRINLLS RVYRPAYAD QEQPPSILDL EKPVDGDIVP VILFFHGGSF AHSSANSAIY DTLCRRLVGL CKCVVVSVNY RRAPENPYPC A YDDGWIAL NWVNSRSWLK SKKDSKVHIF LAGDSSGGNI AHNVALRAGE SGIDVLGNIL LNPMFGGNER TESEKSLDGK YF VTVRDRD WYWKAFLPEG EDREHPACNP FSPRGKSLEG VSFPKSLVVV AGLDLIRDWQ LAYAEGLKKA GQEVKLMHLE KAT VGFYLL PNNNHFHNVM DEISAFVNAE C UniProtKB: Gibberellin receptor GID1A |
-Macromolecule #3: GIBBERELLIN A3
| Macromolecule | Name: GIBBERELLIN A3 / type: ligand / ID: 3 / Number of copies: 1 / Formula: GA3 |
|---|---|
| Molecular weight | Theoretical: 346.374 Da |
| Chemical component information | 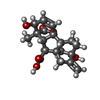 ChemComp-GA3: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 246 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































