[English] 日本語
 Yorodumi
Yorodumi- EMDB-6231: The export receptor Crm1 forms a dimer to promote nuclear export ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6231 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA | |||||||||
 Map data Map data | Random conical tilt reconstruction from negative-stain electron micrographs of a human Crm1/RanGTP dimer bound to the HIV-1 Rev-RRE complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA Nuclear Export / HIV-host interactions / Human Crm1 / Human Ran / HIV-1 Rev / HIV-1 Rev Response Element | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of nuclear pore => GO:0017056 / small GTPase binding => GO:0031267 / cellular response to triglyceride / cellular response to salt / protein binding / HuR (ELAVL1) binds and stabilizes mRNA / annulate lamellae / regulation of proteasomal ubiquitin-dependent protein catabolic process / pre-miRNA export from nucleus / RNA nuclear export complex ...structural constituent of nuclear pore => GO:0017056 / small GTPase binding => GO:0031267 / cellular response to triglyceride / cellular response to salt / protein binding / HuR (ELAVL1) binds and stabilizes mRNA / annulate lamellae / regulation of proteasomal ubiquitin-dependent protein catabolic process / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / nuclear export signal receptor activity / regulation of centrosome duplication / manchette / cellular response to mineralocorticoid stimulus / Regulation of cholesterol biosynthesis by SREBP (SREBF) / host cell nucleolus / importin-alpha family protein binding / regulation of protein export from nucleus / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / protein localization to nucleolus / NEP/NS2 Interacts with the Cellular Export Machinery / tRNA processing in the nucleus / GTP metabolic process / Postmitotic nuclear pore complex (NPC) reformation / Integration of viral DNA into host genomic DNA / Autointegration results in viral DNA circles / nucleocytoplasmic transport / MicroRNA (miRNA) biogenesis / DNA metabolic process / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / 2-LTR circle formation / Vpr-mediated nuclear import of PICs / Early Phase of HIV Life Cycle / Maturation of hRSV A proteins / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / dynein intermediate chain binding / mitotic sister chromatid segregation / ribosomal large subunit export from nucleus / spermatid development / Estrogen-dependent nuclear events downstream of ESR-membrane signaling / protein localization to nucleus / viral process / Binding and entry of HIV virion / positive regulation of protein binding / mRNA transport / sperm flagellum / nuclear pore / mRNA export from nucleus / Cajal body / Cyclin A/B1/B2 associated events during G2/M transition / ribosomal subunit export from nucleus / ribosomal small subunit export from nucleus / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / viral life cycle / NPAS4 regulates expression of target genes / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Transcriptional and post-translational regulation of MITF-M expression and activity / centriole / protein export from nucleus / Resolution of Sister Chromatid Cohesion / Downregulation of TGF-beta receptor signaling / mitotic spindle organization / male germ cell nucleus / hippocampus development / Deactivation of the beta-catenin transactivating complex / RHO GTPases Activate Formins / Assembly Of The HIV Virion / Heme signaling / Transcriptional regulation by small RNAs / Budding and maturation of HIV virion / MAPK6/MAPK4 signaling / recycling endosome / kinetochore / positive regulation of protein import into nucleus / small GTPase binding / protein import into nucleus / Separation of Sister Chromatids / GDP binding / nuclear envelope / melanosome / mitotic cell cycle / ribosome biogenesis / G protein activity / actin cytoskeleton organization / midbody / nuclear membrane / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / DNA-binding transcription factor binding / host cell cytoplasm / cadherin binding / response to xenobiotic stimulus / DNA-binding transcription factor activity / ribonucleoprotein complex / protein heterodimerization activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Booth DS / Cheng Y / Frankel AD | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Authors: David S Booth / Yifan Cheng / Alan D Frankel /  Abstract: The HIV Rev protein routes viral RNAs containing the Rev Response Element (RRE) through the Crm1 nuclear export pathway to the cytoplasm where viral proteins are expressed and genomic RNA is ...The HIV Rev protein routes viral RNAs containing the Rev Response Element (RRE) through the Crm1 nuclear export pathway to the cytoplasm where viral proteins are expressed and genomic RNA is delivered to assembling virions. The RRE assembles a Rev oligomer that displays nuclear export sequences (NESs) for recognition by the Crm1-Ran(GTP) nuclear receptor complex. Here we provide the first view of an assembled HIV-host nuclear export complex using single-particle electron microscopy. Unexpectedly, Crm1 forms a dimer with an extensive interface that enhances association with Rev-RRE and poises NES binding sites to interact with a Rev oligomer. The interface between Crm1 monomers explains differences between Crm1 orthologs that alter nuclear export and determine cellular tropism for viral replication. The arrangement of the export complex identifies a novel binding surface to possibly target an HIV inhibitor and may point to a broader role for Crm1 dimerization in regulating host gene expression. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6231.map.gz emd_6231.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6231-v30.xml emd-6231-v30.xml emd-6231.xml emd-6231.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6231.gif 400_6231.gif 80_6231.gif 80_6231.gif | 28.2 KB 3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6231 http://ftp.pdbj.org/pub/emdb/structures/EMD-6231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6231 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6231.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6231.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Random conical tilt reconstruction from negative-stain electron micrographs of a human Crm1/RanGTP dimer bound to the HIV-1 Rev-RRE complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
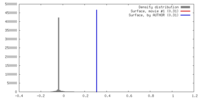
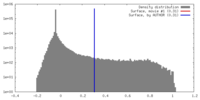
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : A dimer of human Crm1/RanGTP bound to the HIV Rev-RRE complex
| Entire | Name: A dimer of human Crm1/RanGTP bound to the HIV Rev-RRE complex |
|---|---|
| Components |
|
-Supramolecule #1000: A dimer of human Crm1/RanGTP bound to the HIV Rev-RRE complex
| Supramolecule | Name: A dimer of human Crm1/RanGTP bound to the HIV Rev-RRE complex type: sample / ID: 1000 Oligomeric state: Crm1/RanGTP dimer, HIV-1 Rev homohexamer, HIV-1 RRE monomer Number unique components: 4 |
|---|---|
| Molecular weight | Experimental: 462 KDa / Theoretical: 457 KDa / Method: Multi-angle laser light scattering |
-Macromolecule #1: Crm1
| Macromolecule | Name: Crm1 / type: protein_or_peptide / ID: 1 / Name.synonym: Xpo1 / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: Nucleus, cytoplasm Homo sapiens (human) / synonym: Human / Organelle: Nucleus, cytoplasm |
| Molecular weight | Experimental: 125 KDa / Theoretical: 124 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Exportin-1 GO: structural constituent of nuclear pore => GO:0017056, viral process, viral life cycle, small GTPase binding => GO:0031267 InterPro: Armadillo-type fold, Armadillo-like helical, Importin-beta, N-terminal domain, Exportin-1/Importin-beta-like, Exportin-1, C-terminal |
-Macromolecule #2: Ran
| Macromolecule | Name: Ran / type: protein_or_peptide / ID: 2 Details: Mutation of Gln69Leu with guanosine tri-phosphate bound. Number of copies: 2 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Organelle: Nucleus, cytoplasm Homo sapiens (human) / synonym: Human / Organelle: Nucleus, cytoplasm |
| Molecular weight | Experimental: 26 KDa / Theoretical: 25 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: GTP-binding nuclear protein Ran GO: GTP binding, structural constituent of nuclear pore => GO:0017056, viral process, viral life cycle InterPro: P-loop containing nucleoside triphosphate hydrolase, Ran GTPase, Small GTP-binding protein domain, Small GTPase |
-Macromolecule #3: Rev
| Macromolecule | Name: Rev / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Oligomeric state: Hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: HXB3 / synonym: HIV-1 / Organelle: Nucleus, cytoplasm of host human cells Human immunodeficiency virus 1 / Strain: HXB3 / synonym: HIV-1 / Organelle: Nucleus, cytoplasm of host human cells |
| Molecular weight | Theoretical: 13 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Protein Rev GO: viral process, viral life cycle, mRNA transport, RNA binding, host cell nucleus, host cell cytoplasm, protein binding InterPro: Anti-repression trans-activator protein, REV protein |
-Macromolecule #4: Rev Response Element
| Macromolecule | Name: Rev Response Element / type: rna / ID: 4 / Name.synonym: RRE Details: 245-nucleotide portion of the 351-nucleotide Rev Response Element from the HIV-1 genome Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 / Strain: SF-2 / synonym: HIV-1 Human immunodeficiency virus 1 / Strain: SF-2 / synonym: HIV-1 |
| Molecular weight | Theoretical: 80 KDa |
| Sequence | String: GGGCUAGUAG GAGCUAUGUU CCUUGGGUUC UUGGGAGCAG CAGGAAGCAC UAUGGGCGCA GUGUCAUUGA CGCUGACGGU ACAGGCCAGA CAAUUAUUGU CUGGUAUAGU GCAACAGCAG AACAAUUUGC UGAGGGCUAU UGAGGCGCAA CAACAUCUGU UGCAACUCAC ...String: GGGCUAGUAG GAGCUAUGUU CCUUGGGUUC UUGGGAGCAG CAGGAAGCAC UAUGGGCGCA GUGUCAUUGA CGCUGACGGU ACAGGCCAGA CAAUUAUUGU CUGGUAUAGU GCAACAGCAG AACAAUUUGC UGAGGGCUAU UGAGGCGCAA CAACAUCUGU UGCAACUCAC AGUCUGGGGC AUCAAGCAGC UCCAGGCAAG AGUCCUGGCU GUGGAAAGAU ACCUAAGGGA UCAACAGCUC CUAGGGG |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM HEPES-KOH, 50 mM potassium chloride, 2 mM magnesium acetate, 2 mM 2-mercaptoethanol, 2% v/v glycerol |
| Staining | Type: NEGATIVE / Details: 0.75% w/v uranyl formate |
| Grid | Details: 200 mesh copper grid with thin carbon support, glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Electron beam tilt params: 0 |
| Date | Dec 5, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 44 / Average electron dose: 40 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Tilt angle max: 60 |
- Image processing
Image processing
| Details | Random conical tilt reconstructions were generated from samples tilted to 60 degrees using angular parameters from class averages of untilted particles. The structure was further refined by projection matching using both tilted and untilted particles. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: OTHER / Software - Name: FREALIGN, Spider / Number images used: 845 |
| Final two d classification | Number classes: 5 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | A human Crm1 dimer was extracted from the unit cell of the crystal structure. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)