[English] 日本語
 Yorodumi
Yorodumi- EMDB-6197: Negative stain electron microscopy of JRFL SOSIP liganded with PGT151 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6197 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain electron microscopy of JRFL SOSIP liganded with PGT151 | |||||||||
 Map data Map data | Reconstruction of JRFL SOSIP liganded with PGT151 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Simian-Human immunodeficiency virus / unidentified (others) Simian-Human immunodeficiency virus / unidentified (others) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Guenaga J / de Val N / Ward AB / Wyatt RT | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2015 Journal: PLoS Pathog / Year: 2015Title: Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. Authors: Javier Guenaga / Natalia de Val / Karen Tran / Yu Feng / Karen Satchwell / Andrew B Ward / Richard T Wyatt /  Abstract: The structure of BG505 gp140 SOSIP, a soluble mimic of the native HIV-1 envelope glycoprotein (Env), marks the beginning of new era in Env structure-based immunogen design. Displaying a well-ordered ...The structure of BG505 gp140 SOSIP, a soluble mimic of the native HIV-1 envelope glycoprotein (Env), marks the beginning of new era in Env structure-based immunogen design. Displaying a well-ordered quaternary structure, these subtype A-derived trimers display an excellent antigenic profile, discriminating recognition by broadly neutralizing antibodies (bNAbs) from non-broadly neutralizing antibodies (non-bNAbs), and provide a solid Env-based immunogenic platform starting point. Even with this important advance, obtaining homogeneous well-ordered soluble SOSIP trimers derived from other subtypes remains challenging. Here, we report the "rescue" of homogeneous well-ordered subtype B and C SOSIP trimers from a heterogeneous Env mixture using CD4 binding site-directed (CD4bs) non-bNAbs in a negative-selection purification process. These non-bNAbs recognize the primary receptor CD4bs only on disordered trimers but not on the native Env spike or well-ordered soluble trimers due to steric hindrance. Following negative selection to remove disordered oligomers, we demonstrated recovery of well-ordered, homogeneous trimers by electron microscopy (EM). We obtained 3D EM reconstructions of unliganded trimers, as well as in complex with sCD4, a panel of CD4bs-directed bNAbs, and the cleavage-dependent, trimer-specific bNAb, PGT151. Using bio-layer light interferometry (BLI) we demonstrated that the well-ordered trimers were efficiently recognized by bNAbs and poorly recognized by non-bNAbs, representing soluble mimics of the native viral spike. Biophysical characterization was consistent with the thermostability of a homogeneous species that could be further stabilized by specific bNAbs. This study revealed that Env trimers generate different frequencies of well-ordered versus disordered aberrant trimers even when they are genetically identical. By negatively selecting the native-like well-ordered trimers, we establish a new means to obtain soluble Env mimetics derived from subtypes B and C for expanded use as candidate vaccine immunogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6197.map.gz emd_6197.map.gz | 960.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6197-v30.xml emd-6197-v30.xml emd-6197.xml emd-6197.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6197.gif 400_6197.gif 80_6197.gif 80_6197.gif | 9.1 KB 1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6197 http://ftp.pdbj.org/pub/emdb/structures/EMD-6197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6197 | HTTPS FTP |
-Validation report
| Summary document |  emd_6197_validation.pdf.gz emd_6197_validation.pdf.gz | 77.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6197_full_validation.pdf.gz emd_6197_full_validation.pdf.gz | 76.7 KB | Display | |
| Data in XML |  emd_6197_validation.xml.gz emd_6197_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6197 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6197 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6197 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6197 | HTTPS FTP |
-Related structure data
| Related structure data |  6189C  6190C  6191C  6192C  6193C  6194C  6195C  6196C  6198C  6199C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6197.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6197.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
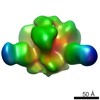
| Annotation | Reconstruction of JRFL SOSIP liganded with PGT151 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : JRFL SOSIP liganded with PGT151
| Entire | Name: JRFL SOSIP liganded with PGT151 |
|---|---|
| Components |
|
-Supramolecule #1000: JRFL SOSIP liganded with PGT151
| Supramolecule | Name: JRFL SOSIP liganded with PGT151 / type: sample / ID: 1000 Oligomeric state: One trimer of JRFL SOSIP binds 2 PGT151 molecules Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 520 KDa / Theoretical: 520 KDa / Method: Size exclusion chromatography (SEC) |
-Macromolecule #1: JRFL SOSIP gp140
| Macromolecule | Name: JRFL SOSIP gp140 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Oligomeric state: trimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Simian-Human immunodeficiency virus Simian-Human immunodeficiency virus |
| Molecular weight | Experimental: 520 KDa / Theoretical: 520 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK 293F Homo sapiens (human) / Recombinant cell: HEK 293F |
-Macromolecule #2: PGT151
| Macromolecule | Name: PGT151 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50 mM Tris-HCl, 150 mM NaCl |
| Staining | Type: NEGATIVE Details: Grids were stained for 30 seconds with 2% uranyl formate. |
| Grid | Details: 400 Cu mesh grids, glow-discharged at 15 mA for 30 seconds |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Date | Feb 7, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 167 / Average electron dose: 39.67 e/Å2 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.0 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 46000 |
| Sample stage | Specimen holder model: OTHER / Tilt angle max: 50 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: EMAN2, sparx / Number images used: 16361 |
|---|
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)