[English] 日本語
 Yorodumi
Yorodumi- EMDB-61685: Cryo-EM structure of the zeaxanthin-bound light-driven proton pum... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the zeaxanthin-bound light-driven proton pumping rhodopsin, NM-R1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | proton pump rhodopsin RETINAL CELL-FREE SYNTHESIS Bacterial type rhodopsin / zeaxanthin / MEMBRANE PROTEIN | |||||||||
| Function / homology | Bacteriorhodopsin-like protein / Archaeal/bacterial/fungal rhodopsins / Bacteriorhodopsin-like protein / photoreceptor activity / phototransduction / membrane / Proteorhodopsin Function and homology information Function and homology information | |||||||||
| Biological species |  Nonlabens marinus S1-08 (bacteria) Nonlabens marinus S1-08 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | |||||||||
 Authors Authors | Hosaka T / Shirouzu M | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2025 Journal: Nat Microbiol / Year: 2025Title: Carotenoids bind rhodopsins and act as photocycle-accelerating pigments in marine Bacteroidota. Authors: Takayoshi Fujiwara / Toshiaki Hosaka / Masumi Hasegawa-Takano / Yosuke Nishimura / Kento Tominaga / Kaho Mori / Satoshi Nishino / Yuno Takahashi / Tomomi Uchikubo-Kamo / Kazuharu Hanada / ...Authors: Takayoshi Fujiwara / Toshiaki Hosaka / Masumi Hasegawa-Takano / Yosuke Nishimura / Kento Tominaga / Kaho Mori / Satoshi Nishino / Yuno Takahashi / Tomomi Uchikubo-Kamo / Kazuharu Hanada / Takashi Maoka / Shinichi Takaichi / Keiichi Inoue / Mikako Shirouzu / Susumu Yoshizawa /  Abstract: Microbial rhodopsins are photoreceptor proteins widely distributed in marine microorganisms that harness light energy and support marine ecosystems. While retinal is typically the sole chromophore in ...Microbial rhodopsins are photoreceptor proteins widely distributed in marine microorganisms that harness light energy and support marine ecosystems. While retinal is typically the sole chromophore in microbial rhodopsins, some proteorhodopsins, which are proton-pumping rhodopsins abundant in the ocean, use carotenoid antennae to transfer light energy to retinal. However, the mechanism by which carotenoids enhance rhodopsin functions remains unclear. Here, using the marine Bacteroidota isolate Nonlabens marinus S1-08, we reconstituted complexes of rhodopsins with the carotenoid myxol and detected energy transfer to retinal in both proteorhodopsin and chloride ion-pumping rhodopsin. Carotenoid binding facilitated light harvesting and accelerated the photocycle, thereby improving the light utilization efficiency of proteorhodopsin. Cryogenic electron microscopy structural analysis further revealed the molecular architecture of the carotenoid-rhodopsin complexes. The ability to bind carotenoids is conserved in rhodopsins of the marine-dominant phylum Bacteroidota, which are widely transcribed in the photic zone. These findings reveal how carotenoids enhance rhodopsin functions in marine Bacteroidota. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61685.map.gz emd_61685.map.gz | 20.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61685-v30.xml emd-61685-v30.xml emd-61685.xml emd-61685.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61685_fsc.xml emd_61685_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_61685.png emd_61685.png | 47.6 KB | ||
| Masks |  emd_61685_msk_1.map emd_61685_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61685.cif.gz emd-61685.cif.gz | 6.2 KB | ||
| Others |  emd_61685_half_map_1.map.gz emd_61685_half_map_1.map.gz emd_61685_half_map_2.map.gz emd_61685_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61685 http://ftp.pdbj.org/pub/emdb/structures/EMD-61685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61685 | HTTPS FTP |
-Related structure data
| Related structure data |  9jouMC  9jovC  9jowC  9joxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61685.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61685.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61685_msk_1.map emd_61685_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61685_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_61685_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NM-R1
| Entire | Name: NM-R1 |
|---|---|
| Components |
|
-Supramolecule #1: NM-R1
| Supramolecule | Name: NM-R1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Nonlabens marinus S1-08 (bacteria) Nonlabens marinus S1-08 (bacteria) |
-Macromolecule #1: Proteorhodopsin
| Macromolecule | Name: Proteorhodopsin / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nonlabens marinus S1-08 (bacteria) Nonlabens marinus S1-08 (bacteria) |
| Molecular weight | Theoretical: 29.481465 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNNLLLNIEV VRLAADDYVG FTFFVGCMAM MAASAFFFLS MSSFERKWRT SILVSGLITF IAAVHYWYMR DYWSGFAESP VFFRYVDWV LTVPLMCVEF YLILKVAGAK KSLMWKLIFL SVVMLVTGYF GEAVDRGNAW LWGLFSGVAY FWIVIEIWFG K AKKLAVAA ...String: MNNLLLNIEV VRLAADDYVG FTFFVGCMAM MAASAFFFLS MSSFERKWRT SILVSGLITF IAAVHYWYMR DYWSGFAESP VFFRYVDWV LTVPLMCVEF YLILKVAGAK KSLMWKLIFL SVVMLVTGYF GEAVDRGNAW LWGLFSGVAY FWIVIEIWFG K AKKLAVAA GGDVLAAHKT LCWFVLVGWA IYPIGYMAGT PGWYDSIFGG WDLNVIYNIG DAINKIGFGL VIYNLAVQAT NK KDGLVNL EENLYFQGWS HPQ UniProtKB: Proteorhodopsin |
-Macromolecule #2: RETINAL
| Macromolecule | Name: RETINAL / type: ligand / ID: 2 / Number of copies: 5 / Formula: RET |
|---|---|
| Molecular weight | Theoretical: 284.436 Da |
| Chemical component information |  ChemComp-RET: |
-Macromolecule #3: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 3 / Number of copies: 13 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Macromolecule #4: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 4 / Number of copies: 12 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Macromolecule #5: Zeaxanthin
| Macromolecule | Name: Zeaxanthin / type: ligand / ID: 5 / Number of copies: 5 / Formula: K3I |
|---|---|
| Molecular weight | Theoretical: 568.871 Da |
| Chemical component information | 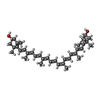 ChemComp-K3I: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 51 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9jou: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)














































