[English] 日本語
 Yorodumi
Yorodumi- EMDB-6167: Essential Structural and Functional Roles of the Cmr4 Subunit in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6167 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex | |||||||||
 Map data Map data | Helical reconstruction of Cmr4-5 filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / Cmr complex / RNA interference / Cmr proteins | |||||||||
| Biological species |   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Ramia NF / Spilman M / Tang L / Shao Y / Elmore J / Hale C / Cocozaki A / Bhattacharya N / Terns RM / Terns MP ...Ramia NF / Spilman M / Tang L / Shao Y / Elmore J / Hale C / Cocozaki A / Bhattacharya N / Terns RM / Terns MP / Li H / Stagg SM | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2014 Journal: Cell Rep / Year: 2014Title: Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Authors: Nancy F Ramia / Michael Spilman / Li Tang / Yaming Shao / Joshua Elmore / Caryn Hale / Alexis Cocozaki / Nilakshee Bhattacharya / Rebecca M Terns / Michael P Terns / Hong Li / Scott M Stagg /  Abstract: The Cmr complex is the multisubunit effector complex of the type III-B clustered regularly interspaced short palindromic repeats (CRISPR)-Cas immune system. The Cmr complex recognizes a target RNA ...The Cmr complex is the multisubunit effector complex of the type III-B clustered regularly interspaced short palindromic repeats (CRISPR)-Cas immune system. The Cmr complex recognizes a target RNA through base pairing with the integral CRISPR RNA (crRNA) and cleaves the target at multiple regularly spaced locations within the complementary region. To understand the molecular basis of the function of this complex, we have assembled information from electron microscopic and X-ray crystallographic structural studies and mutagenesis of a complete Pyrococcus furiosus Cmr complex. Our findings reveal that four helically packed Cmr4 subunits, which make up the backbone of the Cmr complex, act as a platform to support crRNA binding and target RNA cleavage. Interestingly, we found a hook-like structural feature associated with Cmr4 that is likely the site of target RNA binding and cleavage. Our results also elucidate analogies in the mechanisms of crRNA and target molecule binding by the distinct Cmr type III-A and Cascade type I-E complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6167.map.gz emd_6167.map.gz | 831.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6167-v30.xml emd-6167-v30.xml emd-6167.xml emd-6167.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6167_fsc.xml emd_6167_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  400_6167.gif 400_6167.gif 80_6167.gif 80_6167.gif | 27.1 KB 2.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6167 http://ftp.pdbj.org/pub/emdb/structures/EMD-6167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6167 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6167 | HTTPS FTP |
-Validation report
| Summary document |  emd_6167_validation.pdf.gz emd_6167_validation.pdf.gz | 79.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6167_full_validation.pdf.gz emd_6167_full_validation.pdf.gz | 78.7 KB | Display | |
| Data in XML |  emd_6167_validation.xml.gz emd_6167_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6167 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6167 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6167 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6167.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6167.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of Cmr4-5 filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
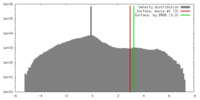
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Filament of Cmr4 and Cmr5
| Entire | Name: Filament of Cmr4 and Cmr5 |
|---|---|
| Components |
|
-Supramolecule #1000: Filament of Cmr4 and Cmr5
| Supramolecule | Name: Filament of Cmr4 and Cmr5 / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Macromolecule #1: Cmr4
| Macromolecule | Name: Cmr4 / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Recombinant expression | Organism:  |
-Macromolecule #2: Cmr5
| Macromolecule | Name: Cmr5 / type: protein_or_peptide / ID: 2 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 20 mM Tris-HCl, pH 7.4, 500 mM NaCl, 5% v/v glycerol, 5 mM beta-mercaptoethanol |
|---|---|
| Grid | Details: 400 mesh Quantifoil grid with 2-micron holes |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | Filaments were prepared for cryoEM by heating a sample of Cmr4-5 to 70 degrees C for 15 minutes and cooling to room temperature over 10 minutes. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 times magnification. Legacy - Electron beam tilt params: 0 |
| Date | Jun 19, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 1866 / Average electron dose: 15 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.3 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)