[English] 日本語
 Yorodumi
Yorodumi- EMDB-61554: Asymmetric structure of cleaved HIV-1 Tri FPPR envelope glycoprot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Asymmetric structure of cleaved HIV-1 Tri FPPR envelope glycoprotein trimer in amphipol-lipid nanodiscs (Tri FPPR.2) | |||||||||
 Map data Map data | The main map with amplitude corrected | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | viral entry / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |  Simian-Human immunodeficiency virus Simian-Human immunodeficiency virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Qi Y / Zhang S / Sodroski J / Mao Y | |||||||||
| Funding support |  China, China,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2025 Journal: Commun Biol / Year: 2025Title: The membrane-proximal external region of human immunodeficiency virus (HIV-1) envelope glycoprotein trimers in A18-lipid nanodiscs. Authors: Yi Qi / Shijian Zhang / Kunyu Wang / Haitao Ding / Zhiqing Zhang / Saumya Anang / Hanh T Nguyen / John C Kappes / Joseph Sodroski / Youdong Mao /   Abstract: During human immunodeficiency virus (HIV-1) entry, the metastable pretriggered envelope glycoprotein (Env) trimer ((gp120/gp41)) opens asymmetrically. We present cryo-EM structures of cleaved ...During human immunodeficiency virus (HIV-1) entry, the metastable pretriggered envelope glycoprotein (Env) trimer ((gp120/gp41)) opens asymmetrically. We present cryo-EM structures of cleaved asymmetric Env trimers in amphipol-lipid nanodiscs. The gp41 membrane-proximal external region (MPER) could be traced in Env protomers that remained close to the nanodisc despite Env tilting. The MPER interacts with the gp120 C-termini and gp41 α9 helices at the base of the Env trimer. MPER conformation is coupled with the tilt angles of the α9 helices, the helicity of the gp41 heptad repeat (HR1) regions, and the opening angles between the protomers of the asymmetric trimers. Our structural models explain the stabilizing effects of MPER integrity and Env proteolytic maturation on the pretriggered Env conformation. Superimposed on the asymmetry of the Env protomers, variation in the glycans at the trimer apex creates substantial structural heterogeneity in the V2 quaternary epitopes of difficult-to-elicit broadly neutralizing antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61554.map.gz emd_61554.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61554-v30.xml emd-61554-v30.xml emd-61554.xml emd-61554.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61554.png emd_61554.png | 133.8 KB | ||
| Filedesc metadata |  emd-61554.cif.gz emd-61554.cif.gz | 6.8 KB | ||
| Others |  emd_61554_half_map_1.map.gz emd_61554_half_map_1.map.gz emd_61554_half_map_2.map.gz emd_61554_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61554 http://ftp.pdbj.org/pub/emdb/structures/EMD-61554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61554 | HTTPS FTP |
-Related structure data
| Related structure data |  9jkgMC  9jkfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61554.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61554.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The main map with amplitude corrected | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_61554_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_61554_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Env Tri FPPR.2
| Entire | Name: HIV-1 Env Tri FPPR.2 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Env Tri FPPR.2
| Supramolecule | Name: HIV-1 Env Tri FPPR.2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Simian-Human immunodeficiency virus Simian-Human immunodeficiency virus |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Simian-Human immunodeficiency virus Simian-Human immunodeficiency virus |
| Molecular weight | Theoretical: 82.23382 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENV TENFNMWKNN MVEQMHEDII SLWDESLKPC VKLTPLCVTL NCTDLRNVTN INNSSEGMRG EIKNCSFNIT T SIRDKVKK ...String: MRVKEKYQHL WRWGWRWGTM LLGMLMICSA TEKLWVTVYY GVPVWKEATT TLFCASDAKA YDTEVHNVWA THACVPTDPN PQEVVLENV TENFNMWKNN MVEQMHEDII SLWDESLKPC VKLTPLCVTL NCTDLRNVTN INNSSEGMRG EIKNCSFNIT T SIRDKVKK DYALFYRLDV VPIDNDNTSY RLINCNTSTI TQACPKVSFE PIPIHYCTPA GFAILKCKDK KFNGTGPCKN VS TVQCTHG IRPVVSTQLL LNGSLAEEEV VIRSSNFTDN AKNIIVQLKE SVEINCTRPN NNTRKSIHIG PGRAFYTTGD IIG DIRQAH CNISRTKWNN TLNQIATKLK EQFGNNKTIV FNQSSGGDPE IVMHSFNCGG EFFYCNSTQL FNSTWNFNGT WNLT QSNGT EGNDTITLPC RIKQIINMWQ EVGKAMYAPP IRGQIRCSSN ITGLILTRDG GNNHNNDTET FRPGGGDMRD NWRSE LYKY KVVKIEPLGV APTKAKRRVV QREKRAVGTI GAMFLGFLGA AGSTMGVASM TLTVQARQLL SGIVQQQNNL LRAIEA QQH LLKLTVWGIK QLQARVLTVE RYLRDQQLLG IWGCSGKLIC TTAVPWNASW SNKTLDMIWN NMTWMEWERE IDNYTGL IY TLIEESQNQQ EKNEQELLEL DKWASLWNWF DITNWLWYIK IFIMIVGGLI GLRIVFTVLS IVNRVRQGYS PGGGHHHH H H UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #13: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 13 / Number of copies: 11 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #14: 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-...
| Macromolecule | Name: 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione type: ligand / ID: 14 / Number of copies: 3 / Formula: 83G |
|---|---|
| Molecular weight | Theoretical: 406.434 Da |
| Chemical component information | 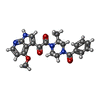 ChemComp-83G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 51.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 50646 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9jkg: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































