[English] 日本語
 Yorodumi
Yorodumi- EMDB-61289: Herpes simplex virus type 1 polymerase machinery in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Herpes simplex virus type 1 polymerase machinery in complex with duplex DNA, acyclovir triphosphate and calcium ions | |||||||||
 Map data Map data | Herpes simplex virus type 1 DNA polymerase machinery in complex with acyclovir triphosphate and calcium ions | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Herpes simplex virus type 1 / HSV-1 / UL30 / UL42 / DNA replication holoenzyme / DNA replication machinery / acyclovir triphosphate / DNA / Calcium / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA polymerase complex / bidirectional double-stranded viral DNA replication / exonuclease activity / DNA polymerase processivity factor activity / DNA-templated DNA replication / RNA-DNA hybrid ribonuclease activity / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding ...DNA polymerase complex / bidirectional double-stranded viral DNA replication / exonuclease activity / DNA polymerase processivity factor activity / DNA-templated DNA replication / RNA-DNA hybrid ribonuclease activity / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / host cell nucleus / DNA binding Similarity search - Function | |||||||||
| Biological species |  Human herpesvirus 1 (strain 17) / synthetic construct (others) Human herpesvirus 1 (strain 17) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Wu YQ / Chen XL / Jiang ZY / Li DY / Zhang ZY / Dong CJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis of herpes simplex type 1 virus DNA polymerase holoenzyme in DNA replication and acyclovir inhibition and proof-reading Authors: Wu YQ / Chen XL / Jiang ZY / Li DY / Zhang ZY / Dong CJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61289.map.gz emd_61289.map.gz | 168.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61289-v30.xml emd-61289-v30.xml emd-61289.xml emd-61289.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_61289.png emd_61289.png | 55.5 KB | ||
| Filedesc metadata |  emd-61289.cif.gz emd-61289.cif.gz | 7.1 KB | ||
| Others |  emd_61289_half_map_1.map.gz emd_61289_half_map_1.map.gz emd_61289_half_map_2.map.gz emd_61289_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61289 http://ftp.pdbj.org/pub/emdb/structures/EMD-61289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61289 | HTTPS FTP |
-Validation report
| Summary document |  emd_61289_validation.pdf.gz emd_61289_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_61289_full_validation.pdf.gz emd_61289_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_61289_validation.xml.gz emd_61289_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_61289_validation.cif.gz emd_61289_validation.cif.gz | 17.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61289 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61289 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61289 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-61289 | HTTPS FTP |
-Related structure data
| Related structure data |  9ja3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61289.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61289.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Herpes simplex virus type 1 DNA polymerase machinery in complex with acyclovir triphosphate and calcium ions | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_61289_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_61289_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Herpes simplex virus type 1 DNA replication machinery in complex ...
| Entire | Name: Herpes simplex virus type 1 DNA replication machinery in complex DNA duplex and dGTP |
|---|---|
| Components |
|
-Supramolecule #1: Herpes simplex virus type 1 DNA replication machinery in complex ...
| Supramolecule | Name: Herpes simplex virus type 1 DNA replication machinery in complex DNA duplex and dGTP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 1 (strain 17) Human herpesvirus 1 (strain 17) |
-Macromolecule #1: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 1 (strain 17) Human herpesvirus 1 (strain 17) |
| Molecular weight | Theoretical: 136.683844 KDa |
| Recombinant expression | Organism: Insect expression vector pBlueBacmsGCA1 (others) |
| Sequence | String: MFSGGGGPLS PGGKSAARAA SGFFAPAGPR GASRGPPPCL RQNFYNPYLA PVGTQQKPTG PTQRHTYYSE CDEFRFIAPR VLDEDAPPE KRAGVHDGHL KRAPKVYCGG DERDVLRVGS GGFWPRRSRL WGGVDHAPAG FNPTVTVFHV YDILENVEHA Y GMRAAQFH ...String: MFSGGGGPLS PGGKSAARAA SGFFAPAGPR GASRGPPPCL RQNFYNPYLA PVGTQQKPTG PTQRHTYYSE CDEFRFIAPR VLDEDAPPE KRAGVHDGHL KRAPKVYCGG DERDVLRVGS GGFWPRRSRL WGGVDHAPAG FNPTVTVFHV YDILENVEHA Y GMRAAQFH ARFMDAITPT GTVITLLGLT PEGHRVAVHV YGTRQYFYMN KEEVDRHLQC RAPRDLCERM AAALRESPGA SF RGISADH FEAEVVERTD VYYYETRPAL FYRVYVRSGR VLSYLCDNFC PAIKKYEGGV DATTRFILDN PGFVTFGWYR LKP GRNNTL AQPRAPMAFG TSSDVEFNCT ADNLAIEGGM SDLPAYKLMC FDIECKAGGE DELAFPVAGH PEDLVIQISC LLYD LSTTA LEHVLLFSLG SCDLPESHLN ELAARGLPTP VVLEFDSEFE MLLAFMTLVK QYGPEFVTGY NIINFDWPFL LAKLT DIYK VPLDGYGRMN GRGVFRVWDI GQSHFQKRSK IKVNGMVNID MYGIITDKIK LSSYKLNAVA EAVLKDKKKD LSYRDI PAY YAAGPAQRGV IGEYCIQDSL LVGQLFFKFL PHLELSAVAR LAGINITRTI YDGQQIRVFT CLLRLADQKG FILPDTQ GR FRGAGGEAPK RPAAAREDEE RPEEEGEDED EREEGGGERE PEGARETAGR HVGYQGARVL DPTSGFHVNP VVVFDFAS L YPSIIQAHNL CFSTLSLRAD AVAHLEAGKD YLEIEVGGRR LFFVKAHVRE SLLSILLRDW LAMRKQIRSR IPQSSPEEA VLLDKQQAAI KVVCNSVYGF TGVQHGLLPC LHVAATVTTI GREMLLATRE YVHARWAAFE QLLADFPEAA DMRAPGPYSM RIIYGDTDS IFVLCRGLTA AGLTAVGDKM ASHISRALFL PPIKLECEKT FTKLLLIAKK KYIGVIYGGK MLIKGVDLVR K NNCAFINR TSRALVDLLF YDDTVSGAAA ALAERPAEEW LARPLPEGLQ AFGAVLVDAH RRITDPERDI QDFVLTAELS RH PRAYTNK RLAHLTVYYK LMARRAQVPS IKDRIPYVIV AQTREVEETV ARLAALRELD AAAPGDEPAP PAALPSPAKR PRE TPSPAD PPGGASKPRK LLVSELAEDP AYAIAHGVAL NTDYYFSHLL GAACVTFKAL FGNNAKITES LLKRFIPEVW HPPD DVAAR LRTAGFGAVG AGATAEETRR MLHRAFDTLA UniProtKB: DNA polymerase |
-Macromolecule #2: DNA polymerase processivity factor
| Macromolecule | Name: DNA polymerase processivity factor / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 1 (strain 17) Human herpesvirus 1 (strain 17) |
| Molecular weight | Theoretical: 54.288227 KDa |
| Recombinant expression | Organism: Insect expression vector pBlueBacmsGCA1His (others) |
| Sequence | String: MHHHHHHHHD YDIPTTENLY FQGADMTDSP GGVAPASPVE DASDASLGQP EEGAPCQVVL QGAELNGILQ AFAPLRTSLL DSLLVMGDR GILIHNTIFG EQVFLPLEHS QFSRYRWRGP TAAFLSLVDQ KRSLLSVFRA NQYPDLRRVE LAITGQAPFR T LVQRIWTT ...String: MHHHHHHHHD YDIPTTENLY FQGADMTDSP GGVAPASPVE DASDASLGQP EEGAPCQVVL QGAELNGILQ AFAPLRTSLL DSLLVMGDR GILIHNTIFG EQVFLPLEHS QFSRYRWRGP TAAFLSLVDQ KRSLLSVFRA NQYPDLRRVE LAITGQAPFR T LVQRIWTT TSDGEAVELA SETLMKRELT SFVVLVPQGT PDVQLRLTRP QLTKVLNATG ADSATPTTFE LGVNGKFSVF TT STCVTFA AREEGVSSST STQVQILSNA LTKAGQAAAN AKTVYGENTH RTFSVVVDDC SMRAVLRRLQ VGGGTLKFFL TTP VPSLCV TATGPNAVSA VFLLKPQKIC LDWLGHSQGS PSAGSSASRA SGSEPTDSQD SASDAVSHGD PEDLDGAARA GEAG ALHAC PMPSSTTRVT PTTKRGRSGG EDARADTALK KPKTGSPTAP PPADPVPLDT EDDSDAADGT AARPAAPDAR SGSRY ACYF RDLPTGEASP GAFSAFRGGP QTPYGFGFP UniProtKB: DNA polymerase processivity factor |
-Macromolecule #3: DNA (27-MER)
| Macromolecule | Name: DNA (27-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.421433 KDa |
| Sequence | String: (DA)(DA)(DT)(DC)(DG)(DT)(DA)(DG)(DG)(DG) (DG)(DA)(DA)(DG)(DG)(DA)(DT)(DC)(DG)(DT) (DA)(DT)(DG)(DG)(DC)(DC)(DT) |
-Macromolecule #4: DNA (5'-D(*AP*GP*GP*CP*CP*AP*TP*AP*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*...
| Macromolecule | Name: DNA (5'-D(*AP*GP*GP*CP*CP*AP*TP*AP*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.921475 KDa |
| Sequence | String: (DA)(DG)(DG)(DC)(DC)(DA)(DT)(DA)(DC)(DG) (DA)(DT)(DC)(DC)(DT)(DT)(DC)(DC)(DC)(DC) (DT)(DA)(DC) |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: ACYCLOVIR TRIPHOSPHATE
| Macromolecule | Name: ACYCLOVIR TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: AVP |
|---|---|
| Molecular weight | Theoretical: 465.144 Da |
| Chemical component information | 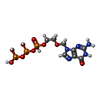 ChemComp-AVP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.4 mg/mL |
|---|---|
| Buffer | pH: 7.2 Details: 25 mM MOPS, pH 7.2, 150mM NaCl, 5% glycerol, 1mM tris(2-carboxyethyl) phosphine (TCEP) |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































