+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6104 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the 80S-RAC complex from yeast | |||||||||
 Map data Map data | Reconstruction of yeast 80S-RAC complex, (Non-rotated state from Dataset RAC5, RAC9, RAC10) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome-associated complex / co-translational chaperone / cryo-em / translation regulation / Zuotin / SSZ | |||||||||
| Biological species |  | |||||||||
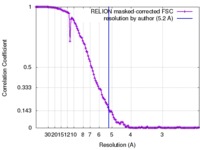
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Zhang YX / Ma CY / Yuan Y / Zhu J / Li NN / Chen C / Wu S / Yu L / Lei JL / Gao N | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2014 Journal: Nat Struct Mol Biol / Year: 2014Title: Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Authors: Yixiao Zhang / Chengying Ma / Yi Yuan / Jing Zhu / Ningning Li / Chu Chen / Shan Wu / Li Yu / Jianlin Lei / Ning Gao /  Abstract: Cotranslational chaperones, ubiquitous in all living organisms, protect nascent polypeptides from aggregation and facilitate their de novo folding. Importantly, emerging data have also suggested that ...Cotranslational chaperones, ubiquitous in all living organisms, protect nascent polypeptides from aggregation and facilitate their de novo folding. Importantly, emerging data have also suggested that ribosome-associated cotranslational chaperones have active regulatory roles in modulating protein translation. By characterizing the structure of a type of eukaryotic cotranslational chaperone, the ribosome-associated complex (RAC) from Saccharomyces cerevisiae, we show that RAC cross-links two ribosomal subunits, through a single long α-helix, to limit the predominant intersubunit rotation required for peptide elongation. We further demonstrate that any changes in the continuity, length or rigidity of this middle α-helix impair RAC function in vivo. Our results suggest a new mechanism in which RAC directly regulates protein translation by mechanically coupling cotranslational folding with the peptide-elongation cycle, and they lay the foundation for further exploration of regulatory roles of RAC in translation control. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6104.map.gz emd_6104.map.gz | 155.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6104-v30.xml emd-6104-v30.xml emd-6104.xml emd-6104.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6104_fsc.xml emd_6104_fsc.xml | 12.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_6104.tif emd_6104.tif | 1.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6104 http://ftp.pdbj.org/pub/emdb/structures/EMD-6104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6104 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6104.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6104.map.gz / Format: CCP4 / Size: 173.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of yeast 80S-RAC complex, (Non-rotated state from Dataset RAC5, RAC9, RAC10) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ribosome-associated complex (RAC) interacting with Saccharomyces ...
| Entire | Name: Ribosome-associated complex (RAC) interacting with Saccharomyces cerevisiae 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: Ribosome-associated complex (RAC) interacting with Saccharomyces ...
| Supramolecule | Name: Ribosome-associated complex (RAC) interacting with Saccharomyces cerevisiae 80S ribosome type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: Saccharomyces cerevisiae 80S ribosome
| Supramolecule | Name: Saccharomyces cerevisiae 80S ribosome / type: complex / ID: 1 / Recombinant expression: Yes / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Ribosome-associated complex
| Macromolecule | Name: Ribosome-associated complex / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 20 mM Hepes-KOH, 120 mM KOAc, 10 mM Mg(OAc)2 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Jan 9, 2013 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Number real images: 8501 / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)