+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of a tmFAP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | de novo protein design / transmembrane protein / ligand binding / fluorogenic / membrane / fluorescent protein. / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Biological species | artificial sequences (others) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Sun K / Zhu JY / Liang MF / Lu PL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: De novo design of transmembrane fluorescence-activating proteins. Authors: Jingyi Zhu / Mingfu Liang / Ke Sun / Yu Wei / Ruiying Guo / Lijing Zhang / Junhui Shi / Dan Ma / Qi Hu / Gaoxingyu Huang / Peilong Lu /  Abstract: The recognition of ligands by transmembrane proteins is essential for the exchange of materials, energy and information across biological membranes. Progress has been made in the de novo design of ...The recognition of ligands by transmembrane proteins is essential for the exchange of materials, energy and information across biological membranes. Progress has been made in the de novo design of transmembrane proteins, as well as in designing water-soluble proteins to bind small molecules, but de novo design of transmembrane proteins that tightly and specifically bind to small molecules remains an outstanding challenge. Here we present the accurate design of ligand-binding transmembrane proteins by integrating deep learning and energy-based methods. We designed pre-organized ligand-binding pockets in high-quality four-helix backbones for a fluorogenic ligand, and generated a transmembrane span using gradient-guided hallucination. The designer transmembrane proteins specifically activated fluorescence of the target fluorophore with mid-nanomolar affinity, exhibiting higher brightness and quantum yield compared to those of enhanced green fluorescent protein. These proteins were highly active in the membrane fraction of live bacterial and eukaryotic cells following expression. The crystal and cryogenic electron microscopy structures of the designer protein-ligand complexes were very close to the structures of the design models. We showed that the interactions between ligands and transmembrane proteins within the membrane can be accurately designed. Our work paves the way for the creation of new functional transmembrane proteins, with a wide range of applications including imaging, ligand sensing and membrane transport. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60929.map.gz emd_60929.map.gz | 25.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60929-v30.xml emd-60929-v30.xml emd-60929.xml emd-60929.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60929.png emd_60929.png | 76.2 KB | ||
| Filedesc metadata |  emd-60929.cif.gz emd-60929.cif.gz | 6.6 KB | ||
| Others |  emd_60929_half_map_1.map.gz emd_60929_half_map_1.map.gz emd_60929_half_map_2.map.gz emd_60929_half_map_2.map.gz | 25.1 MB 25.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60929 http://ftp.pdbj.org/pub/emdb/structures/EMD-60929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60929 | HTTPS FTP |
-Validation report
| Summary document |  emd_60929_validation.pdf.gz emd_60929_validation.pdf.gz | 721.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60929_full_validation.pdf.gz emd_60929_full_validation.pdf.gz | 721.3 KB | Display | |
| Data in XML |  emd_60929_validation.xml.gz emd_60929_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_60929_validation.cif.gz emd_60929_validation.cif.gz | 12.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60929 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60929 | HTTPS FTP |
-Related structure data
| Related structure data |  9ivkMC  8w6eC  8w6fC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60929.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60929.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.14 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60929_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60929_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : De novo design of HBC599 membrane protein binder
| Entire | Name: De novo design of HBC599 membrane protein binder |
|---|---|
| Components |
|
-Supramolecule #1: De novo design of HBC599 membrane protein binder
| Supramolecule | Name: De novo design of HBC599 membrane protein binder / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism: artificial sequences (others) |
-Macromolecule #1: HBC599 membrane protein binder
| Macromolecule | Name: HBC599 membrane protein binder / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: artificial sequences (others) |
| Molecular weight | Theoretical: 33.037648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DEERLKEILF FLLLIIIFVV FLLIVDYKFL EEFKEKNVTD KEEFNEVIKI DEAVMLISAF LLAIAAALLK ELEELIRKRF EEWEKEEET LKKLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKLE DKSPDSPEMK DFRHGFDILV G QIDDALKL ...String: DEERLKEILF FLLLIIIFVV FLLIVDYKFL EEFKEKNVTD KEEFNEVIKI DEAVMLISAF LLAIAAALLK ELEELIRKRF EEWEKEEET LKKLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKLE DKSPDSPEMK DFRHGFDILV G QIDDALKL ANEGKVKEAQ AAAEQLKTTR NAYIQKYLEK AKETMEKRKE IRRELEKILG VLAGLYVGAA FLMVIAKFLT KK IKEENTT DKEKLNEWYE FVYVLLIFAF FIILAIAVVL LKLLEILG |
-Macromolecule #2: Heavy chain, Fab fragment
| Macromolecule | Name: Heavy chain, Fab fragment / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.321084 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVVDFSLHWV RQAPGKGLEW VAYISSSSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARWGYWPGEP WWKAFDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK S |
-Macromolecule #3: Light Chain, Fab fragment
| Macromolecule | Name: Light Chain, Fab fragment / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.353947 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYLYYSLVT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRG |
-Macromolecule #4: 4-[(Z)-1-cyano-2-{6-[(2-hydroxyethyl)(methyl)amino]-1-benzothioph...
| Macromolecule | Name: 4-[(Z)-1-cyano-2-{6-[(2-hydroxyethyl)(methyl)amino]-1-benzothiophen-2-yl}ethenyl]benzonitrile type: ligand / ID: 4 / Number of copies: 1 / Formula: KY6 |
|---|---|
| Molecular weight | Theoretical: 359.444 Da |
| Chemical component information | 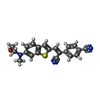 ChemComp-KY6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.74 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 453223 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































