[English] 日本語
 Yorodumi
Yorodumi- EMDB-60904: Structure of SARS-CoV-2 JN.1 spike glycoprotein in complex with A... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SARS-CoV-2 JN.1 spike glycoprotein in complex with ACE2 (2-up state) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | spike protein / glycoprotein / VIRUS / VIRAL PROTEIN / VIRAL PROTEIN-PROTEIN BINDING complex | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
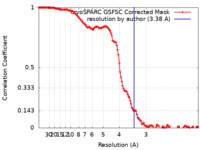
| Method | single particle reconstruction / cryo EM / Resolution: 3.38 Å | ||||||||||||||||||
 Authors Authors | Yajima H / Anraku Y / Kita S / Kimura K / Maenaka K / Hashiguchi T | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1. Authors: Hisano Yajima / Yuki Anraku / Yu Kaku / Kanako Terakado Kimura / Arnon Plianchaisuk / Kaho Okumura / Yoshiko Nakada-Nakura / Yusuke Atarashi / Takuya Hemmi / Daisuke Kuroda / Yoshimasa ...Authors: Hisano Yajima / Yuki Anraku / Yu Kaku / Kanako Terakado Kimura / Arnon Plianchaisuk / Kaho Okumura / Yoshiko Nakada-Nakura / Yusuke Atarashi / Takuya Hemmi / Daisuke Kuroda / Yoshimasa Takahashi / Shunsuke Kita / Jiei Sasaki / Hiromi Sumita / / Jumpei Ito / Katsumi Maenaka / Kei Sato / Takao Hashiguchi /   Abstract: Since 2019, SARS-CoV-2 has undergone mutations, resulting in pandemic and epidemic waves. The SARS-CoV-2 spike protein, crucial for cellular entry, binds to the ACE2 receptor exclusively when its ...Since 2019, SARS-CoV-2 has undergone mutations, resulting in pandemic and epidemic waves. The SARS-CoV-2 spike protein, crucial for cellular entry, binds to the ACE2 receptor exclusively when its receptor-binding domain (RBD) adopts the up-conformation. However, whether ACE2 also interacts with the RBD in the down-conformation to facilitate the conformational shift to RBD-up remains unclear. Herein, we present the structures of the BA.2.86 and the JN.1 spike proteins bound to ACE2. Notably, we successfully observed the ACE2-bound down-RBD, indicating an intermediate structure before the RBD-up conformation. The wider and mobile angle of RBDs in the up-state provides space for ACE2 to interact with the down-RBD, facilitating the transition to the RBD-up state. The K356T, but not N354-linked glycan, contributes to both of infectivity and neutralizing-antibody evasion in BA.2.86. These structural insights the spike-protein dynamics would help understand the mechanisms underlying SARS-CoV-2 infection and its neutralization. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60904.map.gz emd_60904.map.gz | 108 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60904-v30.xml emd-60904-v30.xml emd-60904.xml emd-60904.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60904_fsc.xml emd_60904_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_60904.png emd_60904.png | 94.3 KB | ||
| Masks |  emd_60904_msk_1.map emd_60904_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60904.cif.gz emd-60904.cif.gz | 5.3 KB | ||
| Others |  emd_60904_half_map_1.map.gz emd_60904_half_map_1.map.gz emd_60904_half_map_2.map.gz emd_60904_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60904 http://ftp.pdbj.org/pub/emdb/structures/EMD-60904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60904 | HTTPS FTP |
-Validation report
| Summary document |  emd_60904_validation.pdf.gz emd_60904_validation.pdf.gz | 993.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60904_full_validation.pdf.gz emd_60904_full_validation.pdf.gz | 993.4 KB | Display | |
| Data in XML |  emd_60904_validation.xml.gz emd_60904_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_60904_validation.cif.gz emd_60904_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60904 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60904 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60904 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60904.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60904.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.005 Å | ||||||||||||||||||||||||||||||||||||
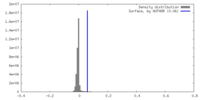
| Density |
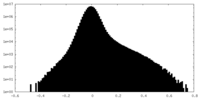
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60904_msk_1.map emd_60904_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
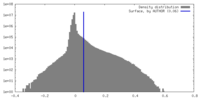
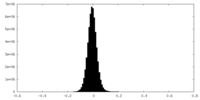
| Density Histograms |
-Half map: #1
| File | emd_60904_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
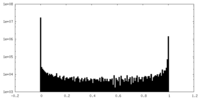
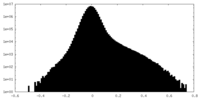
| Density Histograms |
-Half map: #2
| File | emd_60904_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
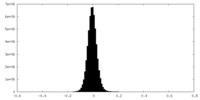
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 JN.1 spike glycoprotein with ACE2 (2-up state)
| Entire | Name: SARS-CoV-2 JN.1 spike glycoprotein with ACE2 (2-up state) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 JN.1 spike glycoprotein with ACE2 (2-up state)
| Supramolecule | Name: SARS-CoV-2 JN.1 spike glycoprotein with ACE2 (2-up state) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: blotting time 5 s and blotting force 5.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 9018 / Average electron dose: 51.233 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)