[English] 日本語
 Yorodumi
Yorodumi- EMDB-60897: Cryo-EM structure of human XPR1 in complex with InsP6 in closed s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human XPR1 in complex with InsP6 in closed state - in the presence of KIDINS220-1-432 without substrate KH2PO4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC53A1 / transporter / phosphate / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphate transmembrane transporter activity / phosphate ion transport / intracellular phosphate ion homeostasis / phosphate ion transmembrane transport / cellular response to phosphate starvation / inositol hexakisphosphate binding / efflux transmembrane transporter activity / response to virus / virus receptor activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zuo P / Liang L / Yin Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate. Authors: Peng Zuo / Weize Wang / Zonglin Dai / Jiye Zheng / Shang Yu / Guangxi Wang / Yue Yin / Ling Liang / Yuxin Yin /  Abstract: Inorganic phosphate (Pi) is essential for life, and its intracellular levels must be tightly regulated to avoid toxicity. XPR1, the sole known phosphate exporter, is critical for maintaining this ...Inorganic phosphate (Pi) is essential for life, and its intracellular levels must be tightly regulated to avoid toxicity. XPR1, the sole known phosphate exporter, is critical for maintaining this balance. Here we report cryo-EM structures of the human XPR1-KIDINS220 complex in substrate-free closed and substrate-bound outward-open states, as well as an XPR1 mutant in a substrate-bound inward-facing state. In the presence of inositol hexaphosphate (InsP) and phosphate, the complex adopts an outward-open conformation, with InsP binding the SPX domain and juxtamembrane regions, indicating active phosphate export. Without phosphate or InsP, the complex closes, with transmembrane helix 9 blocking the outward cavity and a C-terminal loop obstructing the intracellular cavity. XPR1 alone remains closed even with phosphate and InsP. Functional mutagenesis shows that InsP, whose levels vary with Pi availability, works with KIDINS220 to regulate XPR1 activity. These insights into phosphate regulation may aid in developing therapies for ovarian cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60897.map.gz emd_60897.map.gz | 229.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60897-v30.xml emd-60897-v30.xml emd-60897.xml emd-60897.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60897_fsc.xml emd_60897_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_60897.png emd_60897.png | 75.8 KB | ||
| Filedesc metadata |  emd-60897.cif.gz emd-60897.cif.gz | 6.4 KB | ||
| Others |  emd_60897_half_map_1.map.gz emd_60897_half_map_1.map.gz emd_60897_half_map_2.map.gz emd_60897_half_map_2.map.gz | 226.1 MB 226 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60897 http://ftp.pdbj.org/pub/emdb/structures/EMD-60897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60897 | HTTPS FTP |
-Validation report
| Summary document |  emd_60897_validation.pdf.gz emd_60897_validation.pdf.gz | 956.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60897_full_validation.pdf.gz emd_60897_full_validation.pdf.gz | 955.9 KB | Display | |
| Data in XML |  emd_60897_validation.xml.gz emd_60897_validation.xml.gz | 22.2 KB | Display | |
| Data in CIF |  emd_60897_validation.cif.gz emd_60897_validation.cif.gz | 28.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60897 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60897 | HTTPS FTP |
-Related structure data
| Related structure data |  9iucMC  9ineC  9infC  9inhC  9itgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60897.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60897.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : XPR1-KIDINS220-1-432 in complex with InsP6 without substrate KH2PO4
| Entire | Name: XPR1-KIDINS220-1-432 in complex with InsP6 without substrate KH2PO4 |
|---|---|
| Components |
|
-Supramolecule #1: XPR1-KIDINS220-1-432 in complex with InsP6 without substrate KH2PO4
| Supramolecule | Name: XPR1-KIDINS220-1-432 in complex with InsP6 without substrate KH2PO4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 53 member 1
| Macromolecule | Name: Solute carrier family 53 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.673992 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKFAEHLSAH ITPEWRKQYI QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQN ELQSSLDAQK ESTGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI L KKHDKILE ...String: MKFAEHLSAH ITPEWRKQYI QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQN ELQSSLDAQK ESTGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI L KKHDKILE TSRGADWRVA HVEVAPFYTC KKINQLISET EAVVTNELED GDRQKAMKRL RVPPLGAAQP APAWTTFRVG LF CGIFIVL NITLVLAAVF KLETDRSIWP LIRIYRGGFL LIEFLFLLGI NTYGWRQAGV NHVLIFELNP RSNLSHQHLF EIA GFLGIL WCLSLLACFF APISVIPTYV YPLALYGFMV FFLINPTKTF YYKSRFWLLK LLFRVFTAPF HKVGFADFWL ADQL NSLSV ILMDLEYMIC FYSLELKWDE SKGLLPNNSE ESGICHKYTY GVRAIVQCIP AWLRFIQCLR RYRDTKRAFP HLVNA GKYS TTFFMVTFAA LYSTHKERGH SDTMVFFYLW IVFYIISSCY TLIWDLKMDW GLFDKNAGEN TFLREEIVYP QKAYYY CAI IEDVILRFAW TIQISITSTT LLPHSGDIIA TVFAPLEVFR RFVWNFFRLE NEHLNNCGEF RAVRDISVAP LNADDQT LL EQMMDQDDGV RNRQKNRSWK YNQSISLRRP RLASQSKARD TKVLIEDTDD EANTSRENLY FQ UniProtKB: Solute carrier family 53 member 1 |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 12 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Macromolecule #4: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 4 / Number of copies: 6 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 4 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #6: ARACHIDONIC ACID
| Macromolecule | Name: ARACHIDONIC ACID / type: ligand / ID: 6 / Number of copies: 2 / Formula: ACD |
|---|---|
| Molecular weight | Theoretical: 304.467 Da |
| Chemical component information | 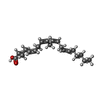 ChemComp-ACD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































