+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human XPR1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC transporter / XPR1 / SLC53A1 / structural protein / Pi / exporter | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphate transmembrane transporter activity / phosphate ion transport / intracellular phosphate ion homeostasis / cellular response to phosphate starvation / phosphate ion transmembrane transport / inositol hexakisphosphate binding / efflux transmembrane transporter activity / response to virus / virus receptor activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | She J / Chen L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structure and function of human XPR1 in phosphate export. Authors: Long Chen / Jin He / Mingxing Wang / Ji She /  Abstract: Xenotropic and polytropic retrovirus receptor 1 (XPR1) functions as a phosphate exporter and is pivotal in maintaining human phosphate homeostasis. It has been identified as a causative gene for ...Xenotropic and polytropic retrovirus receptor 1 (XPR1) functions as a phosphate exporter and is pivotal in maintaining human phosphate homeostasis. It has been identified as a causative gene for primary familial brain calcification. Here we present the cryogenic electron microscopy (cryo-EM) structure of human XPR1 (HsXPR1). HsXPR1 exhibits a dimeric structure in which only TM1 directly constitutes the dimer interface of the transmembrane domain. Each HsXPR1 subunit can be divided spatially into a core domain and a scaffold domain. The core domain of HsXPR1 forms a pore-like structure, along which two phosphate-binding sites enriched with positively charged residues are identified. Mutations of key residues at either site substantially diminish the transport activity of HsXPR1. Phosphate binding at the central site may trigger a conformational change at TM9, leading to the opening of the extracellular gate. In addition, our structural analysis reveals a new conformational state of HsXPR1 in which the cytoplasmic SPX domains form a V-shaped structure. Altogether, our results elucidate the overall architecture of HsXPR1 and shed light on XPR1-mediated phosphate export. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60469.map.gz emd_60469.map.gz | 203.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60469-v30.xml emd-60469-v30.xml emd-60469.xml emd-60469.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60469.png emd_60469.png | 90.9 KB | ||
| Filedesc metadata |  emd-60469.cif.gz emd-60469.cif.gz | 6 KB | ||
| Others |  emd_60469_half_map_1.map.gz emd_60469_half_map_1.map.gz emd_60469_half_map_2.map.gz emd_60469_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60469 http://ftp.pdbj.org/pub/emdb/structures/EMD-60469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60469 | HTTPS FTP |
-Related structure data
| Related structure data |  8ztoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_60469.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60469.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60469_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60469_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The structure of Human XPR1
| Entire | Name: The structure of Human XPR1 |
|---|---|
| Components |
|
-Supramolecule #1: The structure of Human XPR1
| Supramolecule | Name: The structure of Human XPR1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 53 member 1
| Macromolecule | Name: Solute carrier family 53 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.789992 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKFAEHLSAH ITPEWRKQYI QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQN ELQSSLDAQK ESTGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI L KKHDKILE ...String: MKFAEHLSAH ITPEWRKQYI QYEAFKDMLY SAQDQAPSVE VTDEDTVKRY FAKFEEKFFQ TCEKELAKIN TFYSEKLAEA QRRFATLQN ELQSSLDAQK ESTGVTTLRQ RRKPVFHLSH EERVQHRNIK DLKLAFSEFY LSLILLQNYQ NLNFTGFRKI L KKHDKILE TSRGADWRVA HVEVAPFYTC KKINQLISET EAVVTNELED GDRQKAMKRL RVPPLGAAQP APAWTTFRVG LF CGIFIVL NITLVLAAVF KLETDRSIWP LIRIYRGGFL LIEFLFLLGI NTYGWRQAGV NHVLIFELNP RSNLSHQHLF EIA GFLGIL WCLSLLACFF APISVIPTYV YPLALYGFMV FFLINPTKTF YYKSRFWLLK LLFRVFTAPF HKVGFADFWL ADQL NSLSV ILMDLEYMIC FYSLELKWDE SKGLLPNNSE ESGICHKYTY GVRAIVQCIP AWLRFIQCLR RYRDTKRAFP HLVNA GKYS TTFFMVTFAA LYSTHKERGH SDTMVFFYLW IVFYIISSCY TLIWDLKMDW GLFDKNAGEN TFLREEIVYP QKAYYY CAI IEDVILRFAW TIQISITSTT LLPHSGDIIA TVFAPLEVFR RFVWNFFRLE NEHLNNCGEF RAVRDISVAP LNADDQT LL EQMMDQDDGV RNRQKNRSWK YNQSISLRRP RLASQSKARD TKVLIEDTDD EANTASDYKD DDDK UniProtKB: Solute carrier family 53 member 1 |
-Macromolecule #2: [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azany...
| Macromolecule | Name: [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate type: ligand / ID: 2 / Number of copies: 2 / Formula: K9G |
|---|---|
| Molecular weight | Theoretical: 761.084 Da |
| Chemical component information | 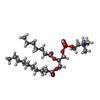 ChemComp-K9G: |
-Macromolecule #3: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































