+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6044 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ribosome conformation along minimum free-energy trajectory | |||||||||
 Map data Map data | Conformation of yeast ribosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Translocation / energy landscape / conformations / Brownian machine | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Dashti A / Schwander P / Langlois R / Fung R / Li W / Hosseinizadeh A / Liao HY / Pallesen J / Sharma G / Stupina VA ...Dashti A / Schwander P / Langlois R / Fung R / Li W / Hosseinizadeh A / Liao HY / Pallesen J / Sharma G / Stupina VA / Simon AE / Dinman J / Frank J / Ourmazd A | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Trajectories of the ribosome as a Brownian nanomachine. Authors: Ali Dashti / Peter Schwander / Robert Langlois / Russell Fung / Wen Li / Ahmad Hosseinizadeh / Hstau Y Liao / Jesper Pallesen / Gyanesh Sharma / Vera A Stupina / Anne E Simon / Jonathan D ...Authors: Ali Dashti / Peter Schwander / Robert Langlois / Russell Fung / Wen Li / Ahmad Hosseinizadeh / Hstau Y Liao / Jesper Pallesen / Gyanesh Sharma / Vera A Stupina / Anne E Simon / Jonathan D Dinman / Joachim Frank / Abbas Ourmazd /  Abstract: A Brownian machine, a tiny device buffeted by the random motions of molecules in the environment, is capable of exploiting these thermal motions for many of the conformational changes in its work ...A Brownian machine, a tiny device buffeted by the random motions of molecules in the environment, is capable of exploiting these thermal motions for many of the conformational changes in its work cycle. Such machines are now thought to be ubiquitous, with the ribosome, a molecular machine responsible for protein synthesis, increasingly regarded as prototypical. Here we present a new analytical approach capable of determining the free-energy landscape and the continuous trajectories of molecular machines from a large number of snapshots obtained by cryogenic electron microscopy. We demonstrate this approach in the context of experimental cryogenic electron microscope images of a large ensemble of nontranslating ribosomes purified from yeast cells. The free-energy landscape is seen to contain a closed path of low energy, along which the ribosome exhibits conformational changes known to be associated with the elongation cycle. Our approach allows model-free quantitative analysis of the degrees of freedom and the energy landscape underlying continuous conformational changes in nanomachines, including those important for biological function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Validation report
| Summary document |  emd_6044_validation.pdf.gz emd_6044_validation.pdf.gz | 78.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6044_full_validation.pdf.gz emd_6044_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_6044_validation.xml.gz emd_6044_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6044 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6044 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6044 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6044.map.gz / Format: CCP4 / Size: 7.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6044.map.gz / Format: CCP4 / Size: 7.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Conformation of yeast ribosome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
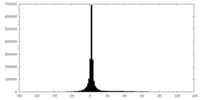
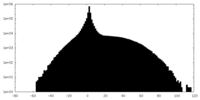
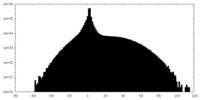
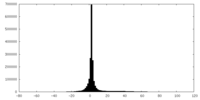
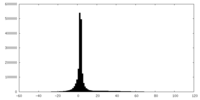
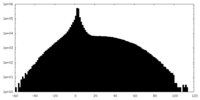
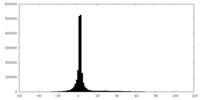
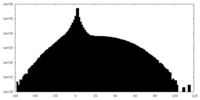
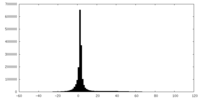
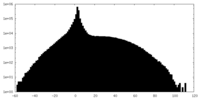
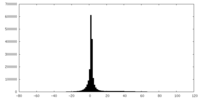
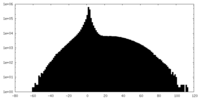
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Supplemental map: emd 6044 additional 1.map
+Supplemental map: emd 6044 additional 10.map
+Supplemental map: emd 6044 additional 11.map
+Supplemental map: emd 6044 additional 12.map
+Supplemental map: emd 6044 additional 13.map
+Supplemental map: emd 6044 additional 14.map
+Supplemental map: emd 6044 additional 15.map
+Supplemental map: emd 6044 additional 16.map
+Supplemental map: emd 6044 additional 17.map
+Supplemental map: emd 6044 additional 18.map
+Supplemental map: emd 6044 additional 19.map
+Supplemental map: emd 6044 additional 2.map
+Supplemental map: emd 6044 additional 20.map
+Supplemental map: emd 6044 additional 21.map
+Supplemental map: emd 6044 additional 22.map
+Supplemental map: emd 6044 additional 23.map
+Supplemental map: emd 6044 additional 24.map
+Supplemental map: emd 6044 additional 25.map
+Supplemental map: emd 6044 additional 26.map
+Supplemental map: emd 6044 additional 27.map
+Supplemental map: emd 6044 additional 28.map
+Supplemental map: emd 6044 additional 29.map
+Supplemental map: emd 6044 additional 3.map
+Supplemental map: emd 6044 additional 30.map
+Supplemental map: emd 6044 additional 31.map
+Supplemental map: emd 6044 additional 32.map
+Supplemental map: emd 6044 additional 33.map
+Supplemental map: emd 6044 additional 34.map
+Supplemental map: emd 6044 additional 35.map
+Supplemental map: emd 6044 additional 36.map
+Supplemental map: emd 6044 additional 37.map
+Supplemental map: emd 6044 additional 38.map
+Supplemental map: emd 6044 additional 39.map
+Supplemental map: emd 6044 additional 4.map
+Supplemental map: emd 6044 additional 40.map
+Supplemental map: emd 6044 additional 41.map
+Supplemental map: emd 6044 additional 42.map
+Supplemental map: emd 6044 additional 43.map
+Supplemental map: emd 6044 additional 44.map
+Supplemental map: emd 6044 additional 45.map
+Supplemental map: emd 6044 additional 46.map
+Supplemental map: emd 6044 additional 47.map
+Supplemental map: emd 6044 additional 48.map
+Supplemental map: emd 6044 additional 49.map
+Supplemental map: emd 6044 additional 5.map
+Supplemental map: emd 6044 additional 6.map
+Supplemental map: emd 6044 additional 7.map
+Supplemental map: emd 6044 additional 8.map
+Supplemental map: emd 6044 additional 9.map
- Sample components
Sample components
-Entire : Yeast 80S ribosome
| Entire | Name: Yeast 80S ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: Yeast 80S ribosome
| Supramolecule | Name: Yeast 80S ribosome / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: 80S ribosome
| Supramolecule | Name: 80S ribosome / type: complex / ID: 1 / Recombinant expression: No / Database: NCBI / Ribosome-details: ribosome-eukaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: carbon-coated Quantifoil 2/4 grid (Quantifoil Micro Tools GmbH, Jena, Germany) |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Jan 1, 2010 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 20 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal magnification: 80000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Number images used: 849914 |
|---|
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)