[English] 日本語
 Yorodumi
Yorodumi- EMDB-60190: Cryo-EM structure of inward-facing Anhydromuropeptide permease (A... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of inward-facing Anhydromuropeptide permease (AmpG) in complex with GlcNAc-1,6-anhMurNAc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AmpG / MFS / Anhydromuropeptide permease / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Yokenella regensburgei (Enteric Group 45) Yokenella regensburgei (Enteric Group 45) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.72 Å | |||||||||
 Authors Authors | Chang N / Kim U / Yoo Y / Kim H / Cho H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural and functional insights of AmpG in muropeptide transport and multiple β-lactam antibiotics resistance. Authors: Nienping Chang / Hoyoung Kim / Uijin Kim / Yongju Cho / Youngki Yoo / Hyunsook Lee / Ji Won Kim / Min Sung Kim / Jaeho Lee / Young-Lag Cho / Kitae Kim / Dongeun Yong / Hyun-Soo Cho /   Abstract: Anhydromuropeptide permease (AmpG) is a transporter protein located in the inner membrane of certain gram -negative bacteria, involved in peptidoglycan (PG) recycling and β-lactamase induction. ...Anhydromuropeptide permease (AmpG) is a transporter protein located in the inner membrane of certain gram -negative bacteria, involved in peptidoglycan (PG) recycling and β-lactamase induction. Decreased AmpG function reduces resistance of antibiotic-resistant bacteria to β-lactam antibiotics. Therefore, AmpG-targeting inhibitors are promising 'antibiotic adjuvants'. However, as the tertiary structure of AmpG has not yet been identified, the development of targeted inhibitors remains challenging. We present four cryo-electron microscopy (cryo-EM) structures: the apo-inward and apo-outward state structures and the inward-occluded and outward states complexed with the substrate GlcNAc-1,6-anhMurNAc. Through functional analysis and molecular dynamics (MD) simulations, we identified motif A, which stabilizes the outward state, substrate-binding pocket, and protonation-related residues. Based on the structure of AmpG and our experimental results, we propose a muropeptide transport mechanism for AmpG. A deeper understanding of its structure and transport mechanism provides a foundation for the development of antibiotic adjuvants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60190.map.gz emd_60190.map.gz | 136.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60190-v30.xml emd-60190-v30.xml emd-60190.xml emd-60190.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_60190.png emd_60190.png | 41.7 KB | ||
| Masks |  emd_60190_msk_1.map emd_60190_msk_1.map | 144.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-60190.cif.gz emd-60190.cif.gz | 7 KB | ||
| Others |  emd_60190_half_map_1.map.gz emd_60190_half_map_1.map.gz emd_60190_half_map_2.map.gz emd_60190_half_map_2.map.gz | 134.3 MB 134.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60190 http://ftp.pdbj.org/pub/emdb/structures/EMD-60190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60190 | HTTPS FTP |
-Related structure data
| Related structure data |  8zkeMC  8zbbC  8zgzC  9j9zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60190.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60190.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.848 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_60190_msk_1.map emd_60190_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_60190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_60190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure ofAnhydromuropeptide permease (AmpG) complex wi...
| Entire | Name: Cryo-EM structure ofAnhydromuropeptide permease (AmpG) complex with GlcNAc-1,6-anhMurNAc |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure ofAnhydromuropeptide permease (AmpG) complex wi...
| Supramolecule | Name: Cryo-EM structure ofAnhydromuropeptide permease (AmpG) complex with GlcNAc-1,6-anhMurNAc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Yokenella regensburgei (Enteric Group 45) Yokenella regensburgei (Enteric Group 45) |
-Macromolecule #1: Muropeptide transporter
| Macromolecule | Name: Muropeptide transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yokenella regensburgei (Enteric Group 45) Yokenella regensburgei (Enteric Group 45) |
| Molecular weight | Theoretical: 54.069086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANHYLRIFQ QPKSAILLIL GFASGLPLAL TSGTLQAWMT VENIDLKTIG FFSLVGQAYV FKFLWSPVMD RYTPPFLGRR RGWLVTTQI LLLIAIAAMG FLEPGTQLRW MAALAVVIAF CSASQDIVFD AWKTDVLPAE ERGTGAAISV LGYRLGMLVS G GLALWMAD ...String: MANHYLRIFQ QPKSAILLIL GFASGLPLAL TSGTLQAWMT VENIDLKTIG FFSLVGQAYV FKFLWSPVMD RYTPPFLGRR RGWLVTTQI LLLIAIAAMG FLEPGTQLRW MAALAVVIAF CSASQDIVFD AWKTDVLPAE ERGTGAAISV LGYRLGMLVS G GLALWMAD KWLGWQGMYW LMAALLVPCI IATLLAPEPS DVVPVPRTLE QAVVAPLRDF FGRNNAWLIL LLIVLYKLGD AF AMSLTTT FLIRGVGFDA GEVGMVNKTL GLIATIIGAL YGGVLMQRLS LFRALLIFGI LQGVSNAGYW LLSITDKHLM SMA VAVFFE NLCGGMGTAA FVALLMTLCN KSFSATQFAL LSALSAVGRV YVGPIAGWFV EAHGWPTFYL FSVFAAVPGI LLLL ICRKT LEYTQQTESF MMRRHFSGAY QFALYLLLLG CLLLALWLIM LALNAIDYTS FSFLAGLLEV AALIAIAGVL LGAIL DYLA LRRTEEHKLA UniProtKB: Anhydromuropeptide permease |
-Macromolecule #2: (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-aceta...
| Macromolecule | Name: (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid type: ligand / ID: 2 / Number of copies: 1 / Formula: 2YP |
|---|---|
| Molecular weight | Theoretical: 478.448 Da |
| Chemical component information | 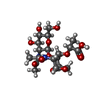 ChemComp-2YP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































