[English] 日本語
 Yorodumi
Yorodumi- EMDB-6016: Dynein motor domain in complex with Lis1 in the presence of ATP and Vi -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6016 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
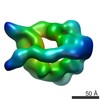
| Title | Dynein motor domain in complex with Lis1 in the presence of ATP and Vi | |||||||||
 Map data Map data | Reconstruction of dynein motor domain in complex with Lis1 in the presence of ATP and Vi | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dynein / lis1 / regulation mechanism | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule sliding / karyogamy / microtubule organizing center organization / nuclear migration along microtubule / astral microtubule / establishment of mitotic spindle localization / microtubule plus-end binding / spindle pole body / vesicle transport along microtubule / minus-end-directed microtubule motor activity ...microtubule sliding / karyogamy / microtubule organizing center organization / nuclear migration along microtubule / astral microtubule / establishment of mitotic spindle localization / microtubule plus-end binding / spindle pole body / vesicle transport along microtubule / minus-end-directed microtubule motor activity / microtubule associated complex / cytoplasmic dynein complex / dynein light intermediate chain binding / nuclear migration / dynein intermediate chain binding / dynein complex binding / Antigen processing: Ubiquitination & Proteasome degradation / mitotic sister chromatid segregation / establishment of mitotic spindle orientation / cytoplasmic microtubule / cytoplasmic microtubule organization / Neutrophil degranulation / mitotic spindle organization / kinetochore / spindle pole / nuclear envelope / cell cortex / cell division / ATP hydrolysis activity / ATP binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 23.1 Å | |||||||||
 Authors Authors | Toropova K / Zou S / Roberts AJ / Redwine WB / Goodman BS / Reck-Peterson SL / Leschziner AE | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: Lis1 regulates dynein by sterically blocking its mechanochemical cycle. Authors: Katerina Toropova / Sirui Zou / Anthony J Roberts / William B Redwine / Brian S Goodman / Samara L Reck-Peterson / Andres E Leschziner /  Abstract: Regulation of cytoplasmic dynein's motor activity is essential for diverse eukaryotic functions, including cell division, intracellular transport, and brain development. The dynein regulator Lis1 is ...Regulation of cytoplasmic dynein's motor activity is essential for diverse eukaryotic functions, including cell division, intracellular transport, and brain development. The dynein regulator Lis1 is known to keep dynein bound to microtubules; however, how this is accomplished mechanistically remains unknown. We have used three-dimensional electron microscopy, single-molecule imaging, biochemistry, and in vivo assays to help establish this mechanism. The three-dimensional structure of the dynein-Lis1 complex shows that binding of Lis1 to dynein's AAA+ ring sterically prevents dynein's main mechanical element, the 'linker', from completing its normal conformational cycle. Single-molecule experiments show that eliminating this block by shortening the linker to a point where it can physically bypass Lis1 renders single dynein motors insensitive to regulation by Lis1. Our data reveal that Lis1 keeps dynein in a persistent microtubule-bound state by directly blocking the progression of its mechanochemical cycle. | |||||||||
| History |
|
- Structure visualization
Structure visualization
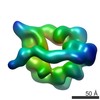
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6016.map.gz emd_6016.map.gz | 17.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6016-v30.xml emd-6016-v30.xml emd-6016.xml emd-6016.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6016.gif 400_6016.gif 80_6016.gif 80_6016.gif | 43.6 KB 3.7 KB | ||
| Others |  emd_6016_half_map_1.map.gz emd_6016_half_map_1.map.gz emd_6016_half_map_2.map.gz emd_6016_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6016 http://ftp.pdbj.org/pub/emdb/structures/EMD-6016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6016 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6016 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6016.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6016.map.gz / Format: CCP4 / Size: 21.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of dynein motor domain in complex with Lis1 in the presence of ATP and Vi | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 6016 half map 1.map
| File | emd_6016_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 6016 half map 2.map
| File | emd_6016_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dynein motor domain in complex with Lis1 in 500 uM ATP/NaVO4
| Entire | Name: Dynein motor domain in complex with Lis1 in 500 uM ATP/NaVO4 |
|---|---|
| Components |
|
-Supramolecule #1000: Dynein motor domain in complex with Lis1 in 500 uM ATP/NaVO4
| Supramolecule | Name: Dynein motor domain in complex with Lis1 in 500 uM ATP/NaVO4 type: sample / ID: 1000 Oligomeric state: One motor domain to one Lis1 propeller domain Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 445 KDa / Theoretical: 445 KDa |
-Macromolecule #1: Dynein heavy chain
| Macromolecule | Name: Dynein heavy chain / type: protein_or_peptide / ID: 1 / Name.synonym: DYN1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 331 KDa / Theoretical: 331 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Dynein heavy chain, cytoplasmic |
-Macromolecule #2: Lis1
| Macromolecule | Name: Lis1 / type: protein_or_peptide / ID: 2 / Name.synonym: Pac1 / Details: Only a single copy of Lis1 is resolved in the map. / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 57 KDa / Theoretical: 57 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Nuclear distribution protein PAC1 |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 50 mM Tris-HCl, 150 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM DTT, 500 uM Mg-ATP/NaVO4 |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein were floated on 2% w/v uranyl formate, then sandwiched with a thin layer of carbon, blotted, and frozen in liquid nitrogen. |
| Grid | Details: 200 mesh C-flat grid with thin carbon support |
| Vitrification | Cryogen name: NITROGEN / Instrument: OTHER Method: Manually blot, wait 10-20 seconds, then manually plunge into liquid nitrogen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Date | Mar 19, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 103 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 86800 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.51 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: phase flip |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 23.1 Å / Resolution method: OTHER / Software - Name: RELION Details: The dataset was first 3D-classified in RELION. Particles in this map displayed a resolved linker domain at lower contour levels. Number images used: 1072 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)