+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | ||||||||||||
 マップデータ マップデータ | |||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | ELMO / DOCK / GEF / GTPASE / RHO / RAC / SIGNALING PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of vascular associated smooth muscle contraction / embryonic olfactory bulb interneuron precursor migration / anatomical structure arrangement / regulation of ERK5 cascade / angiotensin-activated signaling pathway involved in heart process / positive regulation of ovarian follicle development / cerebral cortex GABAergic interneuron development / regulation of respiratory burst / auditory receptor cell morphogenesis / cerebral cortex radially oriented cell migration ...negative regulation of vascular associated smooth muscle contraction / embryonic olfactory bulb interneuron precursor migration / anatomical structure arrangement / regulation of ERK5 cascade / angiotensin-activated signaling pathway involved in heart process / positive regulation of ovarian follicle development / cerebral cortex GABAergic interneuron development / regulation of respiratory burst / auditory receptor cell morphogenesis / cerebral cortex radially oriented cell migration / erythrocyte enucleation / regulation of neutrophil migration / negative regulation of interleukin-23 production / localization within membrane / podosome assembly / Activated NTRK2 signals through CDK5 / interneuron migration / kinocilium / regulation of hydrogen peroxide metabolic process / regulation of cell adhesion involved in heart morphogenesis / negative regulation of receptor-mediated endocytosis / engulfment of apoptotic cell / ruffle assembly / NTRK2 activates RAC1 / Inactivation of CDC42 and RAC1 / NADPH oxidase complex / cochlea morphogenesis / regulation of neuron maturation / respiratory burst / WNT5:FZD7-mediated leishmania damping / SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion / cortical cytoskeleton organization / positive regulation of skeletal muscle acetylcholine-gated channel clustering / hepatocyte growth factor receptor signaling pathway / GTP-dependent protein binding / guanyl-nucleotide exchange factor complex / midbrain dopaminergic neuron differentiation / epithelial cell morphogenesis / cell projection assembly / bone remodeling / positive regulation of bicellular tight junction assembly / ruffle organization / regulation of lamellipodium assembly / thioesterase binding / myoblast fusion / regulation of stress fiber assembly / regulation of neuron migration / negative regulation of fibroblast migration / RHO GTPases activate CIT / cell-cell junction organization / motor neuron axon guidance / positive regulation of vascular associated smooth muscle cell migration / sphingosine-1-phosphate receptor signaling pathway / Nef and signal transduction / PCP/CE pathway / RHO GTPases activate KTN1 / Activation of RAC1 / MET activates RAP1 and RAC1 / regulation of nitric oxide biosynthetic process / DCC mediated attractive signaling / Sema4D mediated inhibition of cell attachment and migration / hyperosmotic response / Azathioprine ADME / Ephrin signaling / CD28 dependent Vav1 pathway / positive regulation of ruffle assembly / positive regulation of neutrophil chemotaxis / positive regulation of cell-substrate adhesion / Wnt signaling pathway, planar cell polarity pathway / superoxide anion generation / podosome / anchoring junction / lamellipodium assembly / regulation of receptor signaling pathway via JAK-STAT / phagocytosis, engulfment / small GTPase-mediated signal transduction / NRAGE signals death through JNK / Activation of RAC1 downstream of NMDARs / dendrite morphogenesis / Rho GDP-dissociation inhibitor binding / regulation of cell size / synaptic transmission, GABAergic / positive regulation of Rho protein signal transduction / positive regulation of dendritic spine development / positive regulation of actin filament polymerization / establishment or maintenance of cell polarity / pericentriolar material / Rac protein signal transduction / RHO GTPases activate PAKs / semaphorin-plexin signaling pathway / positive regulation of epithelial cell migration / ficolin-1-rich granule membrane / regulation of postsynapse assembly / Sema3A PAK dependent Axon repulsion / RHOG GTPase cycle / EPH-ephrin mediated repulsion of cells / regulation of neuronal synaptic plasticity / positive regulation of focal adhesion assembly / RHO GTPases Activate NADPH Oxidases / anatomical structure morphogenesis 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||||||||
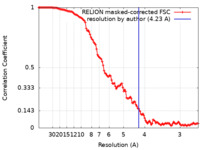
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.23 Å | ||||||||||||
 データ登録者 データ登録者 | Kukimoto-Niino M / Katsura K / Ishizuka-Katsura Y / Mishima-Tsumagari C / Yonemochi M / Inoue M / Nakagawa R / Kaushik R / Zhang KYJ / Shirouzu M | ||||||||||||
| 資金援助 |  日本, 3件 日本, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: J Biol Chem / 年: 2024 ジャーナル: J Biol Chem / 年: 2024タイトル: RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state. 著者: Mutsuko Kukimoto-Niino / Kazushige Katsura / Yoshiko Ishizuka-Katsura / Chiemi Mishima-Tsumagari / Mayumi Yonemochi / Mio Inoue / Reiko Nakagawa / Rahul Kaushik / Kam Y J Zhang / Mikako Shirouzu /  要旨: The dedicator of cytokinesis (DOCK)/engulfment and cell motility (ELMO) complex serves as a guanine nucleotide exchange factor (GEF) for the GTPase Rac. RhoG, another GTPase, activates the ELMO-DOCK- ...The dedicator of cytokinesis (DOCK)/engulfment and cell motility (ELMO) complex serves as a guanine nucleotide exchange factor (GEF) for the GTPase Rac. RhoG, another GTPase, activates the ELMO-DOCK-Rac pathway during engulfment and migration. Recent cryo-EM structures of the DOCK2/ELMO1 and DOCK2/ELMO1/Rac1 complexes have identified closed and open conformations that are key to understanding the autoinhibition mechanism. Nevertheless, the structural details of RhoG-mediated activation of the DOCK/ELMO complex remain elusive. Herein, we present cryo-EM structures of DOCK5/ELMO1 alone and in complex with RhoG and Rac1. The DOCK5/ELMO1 structure exhibits a closed conformation similar to that of DOCK2/ELMO1, suggesting a shared regulatory mechanism of the autoinhibitory state across DOCK-A/B subfamilies (DOCK1-5). Conversely, the RhoG/DOCK5/ELMO1/Rac1 complex adopts an open conformation that differs from that of the DOCK2/ELMO1/Rac1 complex, with RhoG binding to both ELMO1 and DOCK5. The alignment of the DOCK5 phosphatidylinositol (3,4,5)-trisphosphate binding site with the RhoG C-terminal lipidation site suggests simultaneous binding of RhoG and DOCK5/ELMO1 to the plasma membrane. Structural comparison of the apo and RhoG-bound states revealed that RhoG facilitates a closed-to-open state conformational change of DOCK5/ELMO1. Biochemical and surface plasmon resonance (SPR) assays confirm that RhoG enhances the Rac GEF activity of DOCK5/ELMO1 and increases its binding affinity for Rac1. Further analysis of structural variability underscored the conformational flexibility of the DOCK5/ELMO1/Rac1 complex core, potentially facilitating the proximity of the DOCK5 GEF domain to the plasma membrane. These findings elucidate the structural mechanism underlying the RhoG-induced allosteric activation and membrane binding of the DOCK/ELMO complex. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_60148.map.gz emd_60148.map.gz | 140.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-60148-v30.xml emd-60148-v30.xml emd-60148.xml emd-60148.xml | 22.6 KB 22.6 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_60148_fsc.xml emd_60148_fsc.xml | 12 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_60148.png emd_60148.png | 71.2 KB | ||
| Filedesc metadata |  emd-60148.cif.gz emd-60148.cif.gz | 7.7 KB | ||
| その他 |  emd_60148_half_map_1.map.gz emd_60148_half_map_1.map.gz emd_60148_half_map_2.map.gz emd_60148_half_map_2.map.gz | 116.9 MB 117 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60148 http://ftp.pdbj.org/pub/emdb/structures/EMD-60148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60148 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_60148_validation.pdf.gz emd_60148_validation.pdf.gz | 976.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_60148_full_validation.pdf.gz emd_60148_full_validation.pdf.gz | 976.3 KB | 表示 | |
| XML形式データ |  emd_60148_validation.xml.gz emd_60148_validation.xml.gz | 19.3 KB | 表示 | |
| CIF形式データ |  emd_60148_validation.cif.gz emd_60148_validation.cif.gz | 25.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60148 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60148 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8zjkMC  8jhkC  8xm7C  8zj2C  8zjiC  8zjjC  8zjlC  8zjmC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_60148.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_60148.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #1
| ファイル | emd_60148_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_60148_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : DOCK5/ELMO1/Rac1 complex
| 全体 | 名称: DOCK5/ELMO1/Rac1 complex |
|---|---|
| 要素 |
|
-超分子 #1: DOCK5/ELMO1/Rac1 complex
| 超分子 | 名称: DOCK5/ELMO1/Rac1 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Engulfment and cell motility protein 1
| 分子 | 名称: Engulfment and cell motility protein 1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 84.337719 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: GGSGGSMPPP ADIVKVAIEW PGAYPKLMEI DQKKPLSAII KEVCDGWSLA NHEYFALQHA DSSNFYITEK NRNEIKNGTI LRLTTSPAQ NAQQLHERIQ SSSMDAKLEA LKDLASLSRD VTFAQEFINL DGISLLTQMV ESGTERYQKL QKIMKPCFGD M LSFTLTAF ...文字列: GGSGGSMPPP ADIVKVAIEW PGAYPKLMEI DQKKPLSAII KEVCDGWSLA NHEYFALQHA DSSNFYITEK NRNEIKNGTI LRLTTSPAQ NAQQLHERIQ SSSMDAKLEA LKDLASLSRD VTFAQEFINL DGISLLTQMV ESGTERYQKL QKIMKPCFGD M LSFTLTAF VELMDHGIVS WDTFSVAFIK KIASFVNKSA IDISILQRSL AILESMVLNS HDLYQKVAQE ITIGQLIPHL QG SDQEIQT YTIAVINALF LKAPDERRQE MANILAQKQL RSIILTHVIR AQRAINNEMA HQLYVLQVLT FNLLEDRMMT KMD PQDQAQ RDIIFELRRI AFDAESEPNN SSGSMEKRKS MYTRDYKKLG FINHVNPAMD FTQTPPGMLA LDNMLYFAKH HQDA YIRIV LENSSREDKH ECPFGRSSIE LTKMLCEILK VGELPSETCN DFHPMFFTHD RSFEEFFCIC IQLLNKTWKE MRATS EDFN KVMQVVKEQV MRALTTKPSS LDQFKSKLQN LSYTEILKIR QSERMNQEDF QSRPILELKE KIQPEILELI KQQRLN RLV EGTCFRKLNA RRRQDKFWYC RLSPNHKVLH YGDLEESPQG EVPHDSLQDK LPVADIKAVV TGKDCPHMKE KGALKQN KE VLELAFSILY DSNCQLNFIA PDKHEYCIWT DGLNALLGKD MMSDLTRNDL DTLLSMEIKL RLLDLENIQI PDAPPPIP K EPSNYDFVYD CN UniProtKB: Engulfment and cell motility protein 1 |
-分子 #2: Dedicator of cytokinesis protein 5
| 分子 | 名称: Dedicator of cytokinesis protein 5 / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 191.492125 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: GGSGGSMARW IPTKRQKYGV AIYNYNASQD VELSLQIGDT VHILEMYEGW YRGYTLQNKS KKGIFPETYI HLKEATVEDL GQHETVIPG ELPLVQELTS TLREWAVIWR KLYVNNKLTL FRQLQQMTYS LIEWRSQILS GTLPKDELAE LKKKVTAKID H GNRMLGLD ...文字列: GGSGGSMARW IPTKRQKYGV AIYNYNASQD VELSLQIGDT VHILEMYEGW YRGYTLQNKS KKGIFPETYI HLKEATVEDL GQHETVIPG ELPLVQELTS TLREWAVIWR KLYVNNKLTL FRQLQQMTYS LIEWRSQILS GTLPKDELAE LKKKVTAKID H GNRMLGLD LVVRDDNGNI LDPDETSTIA LFKAHEVASK RIEEKIQEEK SILQNLDLRG QSIFSTIHTY GLYVNFKNFV CN IGEDAEL FMALYDPDQS TFISENYLIR WGSNGMPKEI EKLNNLQAVF TDLSSMDLIR PRVSLVCQIV RVGHMELKEG KKH TCGLRR PFGVAVMDIT DIIHGKVDDE EKQHFIPFQQ IAMETYIRQR QLIMSPLITS HVIGENEPLT SVLNKVIAAK EVNH KGQGL WVSLKLLPGD LTQVQKNFSH LVDRSTAIAR KMGFPEIILP GDVRNDIYVT LIHGEFDKGK KKTPKNVEVT MSVHD EEGK LLEKAIHPGA GYEGISEYKS VVYYQVKQPC WYETVKVSIA IEEVTRCHIR FTFRHRSSQE TRDKSERAFG VAFVKL MNP DGTTLQDGRH DLVVYKGDNK KMEDAKFYLT LPGTKMEMEE KELQASKNLV TFTPSKDSTK DSFQIATLIC STKLTQN VD LLGLLNWRSN SQNIKHNLKK LMEVDGGEIV KFLQDTLDAL FNIMMEMSDS ETYDFLVFDA LVFIISLIGD IKFQHFNP V LETYIYKHFS ATLAYVKLSK VLNFYVANAD DSSKTELLFA ALKALKYLFR FIIQSRVLYL RFYGQSKDGD EFNNSIRQL FLAFNMLMDR PLEEAVKIKG AALKYLPSII NDVKLVFDPV ELSVLFCKFI QSIPDNQLVR QKLNCMTKIV ESTLFRQSEC REVLLPLLT DQLSGQLDDN SNKPDHEASS QLLSNILEVL DRKDVGATAV HIQLIMERLL RRINRTVIGM NRQSPHIGSF V ACMIALLQ QMDDSHYSHY ISTFKTRQDI IDFLMETFIM FKDLIGKNVY AKDWMVMNMT QNRVFLRAIN QFAEVLTRFF MD QASFELQ LWNNYFHLAV AFLTHESLQL ETFSQAKRNK IVKKYGDMRK EIGFRIRDMW YNLGPHKIKF IPSMVGPILE VTL TPEVEL RKATIPIFFD MMQCEFNFSG NGNFHMFENE LITKLDQEVE GGRGDEQYKV LLEKLLLEHC RKHKYLSSSG EVFA LLVSS LLENLLDYRT IIMQDESKEN RMSCTVNVLN FYKEKKREDI YIRYLYKLRD LHRDCENYTE AAYTLLLHAE LLQWS DKPC VPHLLQRDSY YVYTQQELKE KLYQEIISYF DKGKMWEKAI KLSKELAETY ESKVFDYEGL GNLLKKRASF YENIIK AMR PQPEYFAVGY YGQGFPSFLR NKIFIYRGKE YERREDFSLR LLTQFPNAEK MTSTTPPGED IKSSPKQYMQ CFTVKPV MS LPPSYKDKPV PEQILNYYRA NEVQQFRYSR PFRKGEKDPD NEFATMWIER TTYTTAYTFP GILKWFEVKQ ISTEEISP L ENAIETMELT NERISNCVQQ HAWDRSLSVH PLSMLLSGIV DPAVMGGFSN YEKAFFTEKY LQEHPEDQEK VELLKRLIA LQMPLLTEGI RIHGEKLTEQ LKPLHERLSS CFRELKEKVE KHYGVITL UniProtKB: Dedicator of cytokinesis protein 5 |
-分子 #3: Ras-related C3 botulinum toxin substrate 1
| 分子 | 名称: Ras-related C3 botulinum toxin substrate 1 / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO / EC番号: small monomeric GTPase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 20.244258 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSSGSSGMQA IKCVVVGDGA VAKTCLLISY TTNAFPGEYI PTVFDNYSAN VMVDGKPVNL GLWDTAGQED YDRLRPLSYP QTDVFLICF SLVSPASFEN VRAKWYPEVR HHCPNTPIIL VGTKLDLRDD KDTIEKLKEK KLTPITYPQG LAMAKEIGAV K YLECSALT QRGLKTVFDE AIRAVL UniProtKB: Ras-related C3 botulinum toxin substrate 1 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 |
|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / メッシュ: 300 / 支持フィルム - #0 - Film type ID: 1 / 支持フィルム - #0 - 材質: CARBON / 支持フィルム - #0 - トポロジー: HOLEY / 支持フィルム - #1 - Film type ID: 2 / 支持フィルム - #1 - 材質: GRAPHENE / 支持フィルム - #1 - トポロジー: HOLEY |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 実像数: 11976 / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.8 µm / 倍率(公称値): 64000 |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)