[English] 日本語
 Yorodumi
Yorodumi- EMDB-5940: 3D single particle reconstruction of the mammalian neuronal nitri... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5940 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D single particle reconstruction of the mammalian neuronal nitric oxide synthase bound to calmodulin | |||||||||
 Map data Map data | Negative-stain EM reconstruction of neuronal nitric oxide synthase bound to calmodulin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nitric oxide synthase / calmodulin / electron transfer | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Yokom AL / Morishima Y / Lau M / Su M / Glukhova A / Osawa Y / Southworth DR | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2014 Journal: J Biol Chem / Year: 2014Title: Architecture of the nitric-oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. Authors: Adam L Yokom / Yoshihiro Morishima / Miranda Lau / Min Su / Alisa Glukhova / Yoichi Osawa / Daniel R Southworth /  Abstract: Nitric-oxide synthase (NOS) is required in mammals to generate NO for regulating blood pressure, synaptic response, and immune defense. NOS is a large homodimer with well characterized reductase and ...Nitric-oxide synthase (NOS) is required in mammals to generate NO for regulating blood pressure, synaptic response, and immune defense. NOS is a large homodimer with well characterized reductase and oxygenase domains that coordinate a multistep, interdomain electron transfer mechanism to oxidize l-arginine and generate NO. Ca(2+)-calmodulin (CaM) binds between the reductase and oxygenase domains to activate NO synthesis. Although NOS has long been proposed to adopt distinct conformations that alternate between interflavin and FMN-heme electron transfer steps, structures of the holoenzyme have remained elusive and the CaM-bound arrangement is unknown. Here we have applied single particle electron microscopy (EM) methods to characterize the full-length of the neuronal isoform (nNOS) complex and determine the structural mechanism of CaM activation. We have identified that nNOS adopts an ensemble of open and closed conformational states and that CaM binding induces a dramatic rearrangement of the reductase domain. Our three-dimensional reconstruction of the intact nNOS-CaM complex reveals a closed conformation and a cross-monomer arrangement with the FMN domain rotated away from the NADPH-FAD center, toward the oxygenase dimer. This work captures, for the first time, the reductase-oxygenase structural arrangement and the CaM-dependent release of the FMN domain that coordinates to drive electron transfer across the domains during catalysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5940.map.gz emd_5940.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5940-v30.xml emd-5940-v30.xml emd-5940.xml emd-5940.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5940.png emd_5940.png | 105.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5940 http://ftp.pdbj.org/pub/emdb/structures/EMD-5940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5940 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5940.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5940.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative-stain EM reconstruction of neuronal nitric oxide synthase bound to calmodulin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
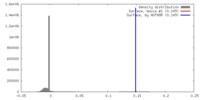
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rat neuronal nitric oxide synthase dimer bound to calmodulin
| Entire | Name: Rat neuronal nitric oxide synthase dimer bound to calmodulin |
|---|---|
| Components |
|
-Supramolecule #1000: Rat neuronal nitric oxide synthase dimer bound to calmodulin
| Supramolecule | Name: Rat neuronal nitric oxide synthase dimer bound to calmodulin type: sample / ID: 1000 Details: The sample was crosslinked with 0.01% glutaraldehyde prior to imaging using negative staining. Oligomeric state: Homodimer of nNOS bound to two CaM proteins Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 360 KDa / Theoretical: 360 KDa / Method: Multi-angle light scattering |
-Macromolecule #1: neuronal nitric oxide synthase
| Macromolecule | Name: neuronal nitric oxide synthase / type: protein_or_peptide / ID: 1 / Name.synonym: nNOS / Number of copies: 2 / Oligomeric state: Homodimer bound to calmodulin / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 360 KDa / Theoretical: 360 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #2: calmodulin
| Macromolecule | Name: calmodulin / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Staining | Type: NEGATIVE / Details: Protein was stained with 0.75% uranyl formate. |
| Grid | Details: 400 mesh copper grids with a thin carbon support |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Date | Jul 10, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 112 / Average electron dose: 30 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Details | Particles were selected manually and 3D refinement was performed using RELION. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: OTHER / Software - Name: EMAN2, RELION, Spider / Number images used: 12323 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)