+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-5830 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Unified assembly mechanism of ASC-dependent inflammasomes | |||||||||
 マップデータ マップデータ | Reconstruction of PYD filament | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | helical polymer / variable twist / apoptosis / death domain | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報NLRP6 inflammasome complex / myosin I binding / Pyrin domain binding / myeloid dendritic cell activation involved in immune response / positive regulation of antigen processing and presentation of peptide antigen via MHC class II / negative regulation of protein serine/threonine kinase activity / regulation of intrinsic apoptotic signaling pathway / myeloid dendritic cell activation / IkappaB kinase complex / The AIM2 inflammasome ...NLRP6 inflammasome complex / myosin I binding / Pyrin domain binding / myeloid dendritic cell activation involved in immune response / positive regulation of antigen processing and presentation of peptide antigen via MHC class II / negative regulation of protein serine/threonine kinase activity / regulation of intrinsic apoptotic signaling pathway / myeloid dendritic cell activation / IkappaB kinase complex / The AIM2 inflammasome / AIM2 inflammasome complex / icosanoid biosynthetic process / NLRP1 inflammasome complex / macropinocytosis / canonical inflammasome complex / interleukin-6 receptor binding / NLRP3 inflammasome complex assembly / BMP receptor binding / positive regulation of adaptive immune response / NLRP3 inflammasome complex / cysteine-type endopeptidase activator activity / CLEC7A/inflammasome pathway / osmosensory signaling pathway / negative regulation of interferon-beta production / regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of extrinsic apoptotic signaling pathway / pattern recognition receptor signaling pathway / positive regulation of macrophage cytokine production / negative regulation of NF-kappaB transcription factor activity / pattern recognition receptor activity / tropomyosin binding / pyroptotic inflammatory response / positive regulation of actin filament polymerization / positive regulation of release of cytochrome c from mitochondria / positive regulation of activated T cell proliferation / intrinsic apoptotic signaling pathway by p53 class mediator / positive regulation of interleukin-10 production / The NLRP3 inflammasome / cellular response to interleukin-1 / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / positive regulation of T cell migration / positive regulation of DNA-binding transcription factor activity / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / positive regulation of defense response to virus by host / negative regulation of canonical NF-kappaB signal transduction / negative regulation of cytokine production involved in inflammatory response / intrinsic apoptotic signaling pathway / activation of innate immune response / positive regulation of phagocytosis / positive regulation of interleukin-1 beta production / tumor necrosis factor-mediated signaling pathway / positive regulation of interleukin-8 production / positive regulation of JNK cascade / apoptotic signaling pathway / positive regulation of non-canonical NF-kappaB signal transduction / positive regulation of NF-kappaB transcription factor activity / protein homooligomerization / positive regulation of T cell activation / regulation of protein stability / positive regulation of interleukin-6 production / positive regulation of type II interferon production / positive regulation of inflammatory response / positive regulation of tumor necrosis factor production / SARS-CoV-1 activates/modulates innate immune responses / cellular response to tumor necrosis factor / azurophil granule lumen / cellular response to lipopolysaccharide / regulation of inflammatory response / protease binding / secretory granule lumen / defense response to Gram-negative bacterium / defense response to virus / microtubule / transmembrane transporter binding / positive regulation of ERK1 and ERK2 cascade / positive regulation of canonical NF-kappaB signal transduction / protein dimerization activity / defense response to Gram-positive bacterium / regulation of autophagy / positive regulation of apoptotic process / inflammatory response / Golgi membrane / innate immune response / neuronal cell body / apoptotic process / Neutrophil degranulation / nucleolus / enzyme binding / endoplasmic reticulum / signal transduction / protein homodimerization activity / protein-containing complex / mitochondrion / extracellular region / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | らせん対称体再構成法 / クライオ電子顕微鏡法 / 解像度: 3.8 Å | |||||||||
 データ登録者 データ登録者 | Lu A / Magupalli VG / Ruan J / Yin Q / Atianand MK / Vos M / Schroder GF / Fitzgerald KA / Wu H / Egelman EH | |||||||||
 引用 引用 |  ジャーナル: Cell / 年: 2014 ジャーナル: Cell / 年: 2014タイトル: Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. 著者: Alvin Lu / Venkat Giri Magupalli / Jianbin Ruan / Qian Yin / Maninjay K Atianand / Matthijn R Vos / Gunnar F Schröder / Katherine A Fitzgerald / Hao Wu / Edward H Egelman /    要旨: Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. ...Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. The adaptor protein ASC bridges the sensor proteins and caspase-1 to form ternary inflammasome complexes, achieved through pyrin domain (PYD) interactions between sensors and ASC and through caspase activation and recruitment domain (CARD) interactions between ASC and caspase-1. We found that PYD and CARD both form filaments. Activated AIM2 and NLRP3 nucleate PYD filaments of ASC, which, in turn, cluster the CARD of ASC. ASC thus nucleates CARD filaments of caspase-1, leading to proximity-induced activation. Endogenous NLRP3 inflammasome is also filamentous. The cryoelectron microscopy structure of ASC(PYD) filament at near-atomic resolution provides a template for homo- and hetero-PYD/PYD associations, as confirmed by structure-guided mutagenesis. We propose that ASC-dependent inflammasomes in both families share a unified assembly mechanism that involves two successive steps of nucleation-induced polymerization. PAPERFLICK: | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_5830.map.gz emd_5830.map.gz | 3.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-5830-v30.xml emd-5830-v30.xml emd-5830.xml emd-5830.xml | 9.1 KB 9.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  400_5830.gif 400_5830.gif 80_5830.gif 80_5830.gif | 76.8 KB 5.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5830 http://ftp.pdbj.org/pub/emdb/structures/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5830 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_5830_validation.pdf.gz emd_5830_validation.pdf.gz | 362.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_5830_full_validation.pdf.gz emd_5830_full_validation.pdf.gz | 361.7 KB | 表示 | |
| XML形式データ |  emd_5830_validation.xml.gz emd_5830_validation.xml.gz | 5.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5830 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5830 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_5830.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_5830.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Reconstruction of PYD filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
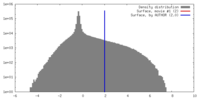
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : PYD domain from ASC
| 全体 | 名称: PYD domain from ASC |
|---|---|
| 要素 |
|
-超分子 #1000: PYD domain from ASC
| 超分子 | 名称: PYD domain from ASC / タイプ: sample / ID: 1000 / 集合状態: helical polymer / Number unique components: 1 |
|---|
-分子 #1: Apoptosis-associated speck-like protein containing a CARD
| 分子 | 名称: Apoptosis-associated speck-like protein containing a CARD タイプ: protein_or_peptide / ID: 1 / Name.synonym: pyrin domain, death domain / 集合状態: helical polymer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) / 別称: Human Homo sapiens (ヒト) / 別称: Human |
| 組換発現 | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | らせん対称体再構成法 |
| 試料の集合状態 | helical array |
- 試料調製
試料調製
| グリッド | 詳細: over holes in lacey carbon grids, no support |
|---|---|
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 日付 | 2012年12月20日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: FEI FALCON II (4k x 4k) 実像数: 400 / 平均電子線量: 20 e/Å2 / 詳細: Integrating mode used |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 5.0 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 詳細 | IHRSR |
|---|---|
| 最終 再構成 | 想定した対称性 - らせんパラメータ - Δz: 13.95 Å 想定した対称性 - らせんパラメータ - ΔΦ: 52.9 ° 想定した対称性 - らせんパラメータ - 軸対称性: C3 (3回回転対称) アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 3.8 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: Spider, IHRSR |
| CTF補正 | 詳細: each image |
 ムービー
ムービー コントローラー
コントローラー





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)