+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5830 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unified assembly mechanism of ASC-dependent inflammasomes | |||||||||
 Map data Map data | Reconstruction of PYD filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | helical polymer / variable twist / apoptosis / death domain | |||||||||
| Function / homology |  Function and homology information Function and homology informationNLRP6 inflammasome complex / myosin I binding / Pyrin domain binding / myeloid dendritic cell activation involved in immune response / positive regulation of antigen processing and presentation of peptide antigen via MHC class II / negative regulation of protein serine/threonine kinase activity / regulation of intrinsic apoptotic signaling pathway / myeloid dendritic cell activation / IkappaB kinase complex / The AIM2 inflammasome ...NLRP6 inflammasome complex / myosin I binding / Pyrin domain binding / myeloid dendritic cell activation involved in immune response / positive regulation of antigen processing and presentation of peptide antigen via MHC class II / negative regulation of protein serine/threonine kinase activity / regulation of intrinsic apoptotic signaling pathway / myeloid dendritic cell activation / IkappaB kinase complex / The AIM2 inflammasome / interleukin-6 receptor binding / AIM2 inflammasome complex / icosanoid biosynthetic process / NLRP1 inflammasome complex / macropinocytosis / canonical inflammasome complex / NLRP3 inflammasome complex assembly / BMP receptor binding / NLRP3 inflammasome complex / positive regulation of adaptive immune response / cysteine-type endopeptidase activator activity / CLEC7A/inflammasome pathway / negative regulation of interferon-beta production / osmosensory signaling pathway / regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of extrinsic apoptotic signaling pathway / pattern recognition receptor signaling pathway / positive regulation of macrophage cytokine production / : / tropomyosin binding / pattern recognition receptor activity / positive regulation of release of cytochrome c from mitochondria / positive regulation of actin filament polymerization / intrinsic apoptotic signaling pathway by p53 class mediator / pyroptotic inflammatory response / positive regulation of activated T cell proliferation / positive regulation of interleukin-10 production / The NLRP3 inflammasome / cellular response to interleukin-1 / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / positive regulation of T cell migration / : / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / positive regulation of defense response to virus by host / negative regulation of cytokine production involved in inflammatory response / activation of innate immune response / intrinsic apoptotic signaling pathway / positive regulation of phagocytosis / negative regulation of canonical NF-kappaB signal transduction / tumor necrosis factor-mediated signaling pathway / positive regulation of interleukin-1 beta production / positive regulation of interleukin-8 production / apoptotic signaling pathway / regulation of protein stability / positive regulation of non-canonical NF-kappaB signal transduction / : / protein homooligomerization / regulation of autophagy / positive regulation of JNK cascade / positive regulation of interleukin-6 production / positive regulation of type II interferon production / positive regulation of T cell activation / positive regulation of tumor necrosis factor production / positive regulation of inflammatory response / SARS-CoV-1 activates/modulates innate immune responses / azurophil granule lumen / cellular response to tumor necrosis factor / regulation of inflammatory response / cellular response to lipopolysaccharide / protease binding / secretory granule lumen / defense response to virus / defense response to Gram-negative bacterium / microtubule / transmembrane transporter binding / positive regulation of canonical NF-kappaB signal transduction / positive regulation of ERK1 and ERK2 cascade / protein dimerization activity / defense response to Gram-positive bacterium / positive regulation of apoptotic process / inflammatory response / Golgi membrane / innate immune response / neuronal cell body / apoptotic process / Neutrophil degranulation / nucleolus / enzyme binding / endoplasmic reticulum / signal transduction / protein homodimerization activity / protein-containing complex / mitochondrion / extracellular region / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Lu A / Magupalli VG / Ruan J / Yin Q / Atianand MK / Vos M / Schroder GF / Fitzgerald KA / Wu H / Egelman EH | |||||||||
 Citation Citation |  Journal: Cell / Year: 2014 Journal: Cell / Year: 2014Title: Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Authors: Alvin Lu / Venkat Giri Magupalli / Jianbin Ruan / Qian Yin / Maninjay K Atianand / Matthijn R Vos / Gunnar F Schröder / Katherine A Fitzgerald / Hao Wu / Edward H Egelman /    Abstract: Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. ...Inflammasomes elicit host defense inside cells by activating caspase-1 for cytokine maturation and cell death. AIM2 and NLRP3 are representative sensor proteins in two major families of inflammasomes. The adaptor protein ASC bridges the sensor proteins and caspase-1 to form ternary inflammasome complexes, achieved through pyrin domain (PYD) interactions between sensors and ASC and through caspase activation and recruitment domain (CARD) interactions between ASC and caspase-1. We found that PYD and CARD both form filaments. Activated AIM2 and NLRP3 nucleate PYD filaments of ASC, which, in turn, cluster the CARD of ASC. ASC thus nucleates CARD filaments of caspase-1, leading to proximity-induced activation. Endogenous NLRP3 inflammasome is also filamentous. The cryoelectron microscopy structure of ASC(PYD) filament at near-atomic resolution provides a template for homo- and hetero-PYD/PYD associations, as confirmed by structure-guided mutagenesis. We propose that ASC-dependent inflammasomes in both families share a unified assembly mechanism that involves two successive steps of nucleation-induced polymerization. PAPERFLICK: | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5830.map.gz emd_5830.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5830-v30.xml emd-5830-v30.xml emd-5830.xml emd-5830.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  400_5830.gif 400_5830.gif 80_5830.gif 80_5830.gif | 76.8 KB 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5830 http://ftp.pdbj.org/pub/emdb/structures/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5830 | HTTPS FTP |
-Related structure data
| Related structure data |  3j63MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5830.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5830.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of PYD filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
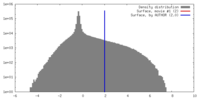
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PYD domain from ASC
| Entire | Name: PYD domain from ASC |
|---|---|
| Components |
|
-Supramolecule #1000: PYD domain from ASC
| Supramolecule | Name: PYD domain from ASC / type: sample / ID: 1000 / Oligomeric state: helical polymer / Number unique components: 1 |
|---|
-Macromolecule #1: Apoptosis-associated speck-like protein containing a CARD
| Macromolecule | Name: Apoptosis-associated speck-like protein containing a CARD type: protein_or_peptide / ID: 1 / Name.synonym: pyrin domain, death domain / Oligomeric state: helical polymer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Grid | Details: over holes in lacey carbon grids, no support |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Dec 20, 2012 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 400 / Average electron dose: 20 e/Å2 / Details: Integrating mode used |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | IHRSR |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 13.95 Å Applied symmetry - Helical parameters - Δ&Phi: 52.9 ° Applied symmetry - Helical parameters - Axial symmetry: C3 (3 fold cyclic) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: OTHER / Software - Name: Spider, IHRSR |
| CTF correction | Details: each image |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)