[English] 日本語
 Yorodumi
Yorodumi- EMDB-53685: Local refinement of the N-terminal domain (NTD) from the Porcine ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of the N-terminal domain (NTD) from the Porcine hemagglutinating encephalomyelitis virus (PHEV) Spike in the open conformation bound to 9-O-Ac-Sia | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coronavirus / Spike / Entry / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Porcine hemagglutinating encephalomyelitis virus Porcine hemagglutinating encephalomyelitis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Fernandez I / Rey FA | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2025 Journal: Nat Microbiol / Year: 2025Title: Dipeptidase 1 is a functional receptor for a porcine coronavirus. Authors: Jérémy Dufloo / Ignacio Fernández / Atousa Arbabian / Ahmed Haouz / Nigel Temperton / Luis G Gimenez-Lirola / Félix A Rey / Rafael Sanjuán /     Abstract: Coronaviruses of the subgenus Embecovirus include several important pathogens, such as the human seasonal coronaviruses HKU1 and OC43, bovine coronavirus and porcine haemagglutinating ...Coronaviruses of the subgenus Embecovirus include several important pathogens, such as the human seasonal coronaviruses HKU1 and OC43, bovine coronavirus and porcine haemagglutinating encephalomyelitis virus (PHEV). While sialic acid is thought to be required for embecovirus entry, protein receptors remain unknown for most of these viruses. Here we show that PHEV does not require sialic acid for entry and instead uses dipeptidase 1 (DPEP1) as a receptor. Cryo-electron microscopy at 3.4-4.4 Å resolution revealed that, unlike other embecoviruses, PHEV displays both open and closed conformations of its spike trimer at steady state. The spike receptor-binding domain (RBD) exhibits extremely high sequence variability across embecoviruses, and we found that DPEP1 usage is specific to PHEV. In contrast, the X-ray structure of the RBD-DPEP1 complex at 2.25 Å showed that the structural elements involved in receptor binding are conserved, highlighting the remarkable versatility of this structural organization in adopting novel receptor specificities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53685.map.gz emd_53685.map.gz | 30.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53685-v30.xml emd-53685-v30.xml emd-53685.xml emd-53685.xml | 31.7 KB 31.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_53685.png emd_53685.png | 40.6 KB | ||
| Masks |  emd_53685_msk_1.map emd_53685_msk_1.map | 61 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-53685.cif.gz emd-53685.cif.gz | 7.5 KB | ||
| Others |  emd_53685_additional_1.map.gz emd_53685_additional_1.map.gz emd_53685_additional_2.map.gz emd_53685_additional_2.map.gz emd_53685_half_map_1.map.gz emd_53685_half_map_1.map.gz emd_53685_half_map_2.map.gz emd_53685_half_map_2.map.gz | 54.4 MB 30.8 MB 56.7 MB 56.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53685 http://ftp.pdbj.org/pub/emdb/structures/EMD-53685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53685 | HTTPS FTP |
-Validation report
| Summary document |  emd_53685_validation.pdf.gz emd_53685_validation.pdf.gz | 713.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_53685_full_validation.pdf.gz emd_53685_full_validation.pdf.gz | 713.3 KB | Display | |
| Data in XML |  emd_53685_validation.xml.gz emd_53685_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  emd_53685_validation.cif.gz emd_53685_validation.cif.gz | 14.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-53685 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-53685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-53685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-53685 | HTTPS FTP |
-Related structure data
| Related structure data |  9r6rMC  9h0bC  9h3jC  9r6oC  9r6pC  9r6qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_53685.map.gz / Format: CCP4 / Size: 61 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53685.map.gz / Format: CCP4 / Size: 61 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_53685_msk_1.map emd_53685_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map sharpened with DeepEMhancer
| File | emd_53685_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map sharpened with DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Consensus map used for the local refinement
| File | emd_53685_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map used for the local refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_53685_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_53685_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Spike
| Entire | Name: Spike |
|---|---|
| Components |
|
-Supramolecule #1: Spike
| Supramolecule | Name: Spike / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Trimeric protein obtained by recombinant expression |
|---|---|
| Source (natural) | Organism:  Porcine hemagglutinating encephalomyelitis virus Porcine hemagglutinating encephalomyelitis virus |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porcine hemagglutinating encephalomyelitis virus Porcine hemagglutinating encephalomyelitis virus |
| Molecular weight | Theoretical: 147.88475 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VLGDLKCNTS SINDVDTGVP SISSEVVDVT NGLGTFYVLD RVYLNTTLLL NGYYPISGAT FRNMALKGTR LLSTLWFKPP FLSPFNDGI FAKVKNSRFF KDGVIYSEFP AITIGSTFVN TSYSIVVEPH TLLINGNLQG LLQISVCQYT MCEYPHTICH P NLGNQRIE ...String: VLGDLKCNTS SINDVDTGVP SISSEVVDVT NGLGTFYVLD RVYLNTTLLL NGYYPISGAT FRNMALKGTR LLSTLWFKPP FLSPFNDGI FAKVKNSRFF KDGVIYSEFP AITIGSTFVN TSYSIVVEPH TLLINGNLQG LLQISVCQYT MCEYPHTICH P NLGNQRIE LWHYDTDVVS CLYRRNFTYD VNADYLYFHF YQEGGTFYAY FTDTGFVTKF LFKLYLGTVL SHYYVMPLTC DS ALSLEYW VTPLTTRQFL LAFDQDGVLY HAVDCASDFM SEIMCKTSSI TPPTGVYELN GYTVQPVATV YRRIPDLPNC DIE AWLNSK TVSSPLNWER KIFSNCNFNM GRLMSFIQAD SFGCNNIDAS RLYGMCFGSI TIDKFAIPNS RKVDLQVGKS GYLQ SFNYK IDTAVSSCQL YYSLPAANVS VTHYNPSSWN RRYGFNNQSF GSRGLHDAVY SQQCFNTPNT YCPCRTSQCI GGAGT GTCP VGTTVRKCFA AVTNATKCTC WCQPDPSTYK GVNAWTCPQS KVSIQPGQHC PGLGLVEDDC SGNPCTCKPQ AFIGWS SET CLQNGRCNIF ANFILNDVNS GTTCSTDLQQ GNTNITTDVC VNYDLYGITG QGILIEVNAT YYNSWQNLLY DSSGNLY GF RDYLSNRTFL IRSCYSGRVS AVFHANSSEP ALMFRNLKCS HVFNNTILRQ IQLVNYFDSY LGCVVNAYNN TASAVSTC D LTVGSGYCVD YVTALRSRRS FTTGYRFTNF EPFAVNLVND SIEPVGGLYE IQIPSEFTIG NLEEFIQTSS PKVTIDCAT FVCGDYAACR QQLAEYGSFC ENINAILIEV NELLDTTQLQ VANSLMNGVT LSTKIKDGIN FNVDDINFSP VLGCLGSECN RASTRSAIE DLLFDKVKLS DVGFVQAYNN CTGGAEIRDL ICVQSYNGIK VLPPLLSENQ ISGYTLAATA ASLFPPWTAA A GVPFYLNV QYRINGLGVT MDVLSQNQKL IASAFNNALD AIQEGFDATN SALVKIQSVV NANAEALNNL LQQLSNRFGA IS ASLQEIL SRLDALEAKA QIDRLINGRL TALNAYVSQQ LSDSTLVKFS AAQAMEKVNE CVKSQSSRIN FCGNGNHIIS LVQ NAPYGL YFIHFSYVPT KYVTAKVSPG LCIAGDIGIS PKSGYFINVN NSWMFTGSGY YYPEPITQNN VVMMSTCAVN YTKA PDLML NTSTPNLPDF KEELYQWFKN QSSVAPDLSL DYINVTFLDL QDEMNRLQEA IKVLNGSGYI PEAPRDGQAY VRKDG EWVL LSTFLGSLVP RGSHHHHHHH HSAWSHPQFE KGTGGLNDIF EAQKIEWHE UniProtKB: Spike glycoprotein |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-...
| Macromolecule | Name: 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid type: ligand / ID: 4 / Number of copies: 1 / Formula: 5N6 |
|---|---|
| Molecular weight | Theoretical: 351.307 Da |
| Chemical component information | 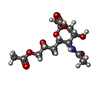 ChemComp-5N6: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: Tris 10 mM, NaCl 100 mM, pH 8.0 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 240000 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




























































