+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5358 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA-packaged Capsid of Bacteriophage phi6 | |||||||||
 Map data Map data | RNA-packaged capsid of the bacteriophage phi6 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dsRNA virus / Cystoviridae / capsid / expansion intermediate / segmented genome | |||||||||
| Biological species |  Bacteriophage phi6 (virus) Bacteriophage phi6 (virus) | |||||||||
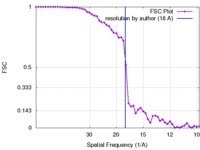
| Method | single particle reconstruction / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Nemecek D / Cheng N / Qiao J / Mindich L / Steven AC / Heymann JB | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2011 Journal: J Mol Biol / Year: 2011Title: Stepwise expansion of the bacteriophage ϕ6 procapsid: possible packaging intermediates. Authors: Daniel Nemecek / Naiqian Cheng / Jian Qiao / Leonard Mindich / Alasdair C Steven / J Bernard Heymann /  Abstract: The initial assembly product of bacteriophage ϕ6, the procapsid, undergoes major structural transformation during the sequential packaging of its three segments of single-stranded RNA. The ...The initial assembly product of bacteriophage ϕ6, the procapsid, undergoes major structural transformation during the sequential packaging of its three segments of single-stranded RNA. The procapsid, a compact icosahedrally symmetric particle with deeply recessed vertices, expands to the spherical mature capsid, increasing the volume available to accommodate the genome by 2.5-fold. It has been proposed that expansion and packaging are linked, with each stage in expansion presenting a binding site for a particular RNA segment. To investigate procapsid transformability, we induced expansion by acidification, heating, and elevated salt concentration. Cryo-electron microscopy reconstructions after all three treatments yielded the same partially expanded particle. Analysis by cryo-electron tomography showed that all vertices of a given capsid were either in a compact or an expanded state, indicating a highly cooperative transition. To benchmark the mature capsid, we analyzed filled (in vivo packaged) capsids. When these particles were induced to release their RNA, they reverted to the same intermediate state as expanded procapsids (intermediate 1) or to a second, further expanded state (intermediate 2). This partial reversibility of expansion suggests that the mature spherical capsid conformation is obtained only when sufficient outward pressure is exerted by packaged RNA. The observation of two intermediates is consistent with the proposed three-step packaging process. The model is further supported by the observation that a mutant capable of packaging the second RNA segment without previously packaging the first segment has enhanced susceptibility for switching spontaneously from the procapsid to the first intermediate state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5358.map.gz emd_5358.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5358-v30.xml emd-5358-v30.xml emd-5358.xml emd-5358.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_5358_fsc.xml emd_5358_fsc.xml | 4.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_5358_1.jpg emd_5358_1.jpg | 70.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5358 http://ftp.pdbj.org/pub/emdb/structures/EMD-5358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5358 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5358 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5358.map.gz / Format: CCP4 / Size: 65.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5358.map.gz / Format: CCP4 / Size: 65.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNA-packaged capsid of the bacteriophage phi6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62792 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RNA-packaged P1247 Capsid of Bacteriophage phi6
| Entire | Name: RNA-packaged P1247 Capsid of Bacteriophage phi6 |
|---|---|
| Components |
|
-Supramolecule #1000: RNA-packaged P1247 Capsid of Bacteriophage phi6
| Supramolecule | Name: RNA-packaged P1247 Capsid of Bacteriophage phi6 / type: sample / ID: 1000 Oligomeric state: Icosahedral shell of P1 dimers with P2, P4 and P7 subunits and packaged RNA Number unique components: 5 |
|---|---|
| Molecular weight | Theoretical: 12.6 MDa |
-Supramolecule #1: Bacteriophage phi6
| Supramolecule | Name: Bacteriophage phi6 / type: virus / ID: 1 / Name.synonym: Bacteriophage phi6 / Sci species name: Bacteriophage phi6 / Database: NCBI / Virus type: OTHER / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes / Syn species name: Bacteriophage phi6 |
|---|---|
| Host (natural) | Organism:  Pseudomonas syringae (bacteria) / synonym: BACTERIA(EUBACTERIA) Pseudomonas syringae (bacteria) / synonym: BACTERIA(EUBACTERIA) |
| Molecular weight | Theoretical: 12.6 MDa |
| Virus shell | Shell ID: 1 / Name: P1 / Diameter: 440 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 10 mM potassium phosphate, 1 mM MgCl2, 100 mM NaCl |
| Grid | Details: Quantifoil 2/2/400 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 93 K / Instrument: LEICA KF80 / Details: Vitrification instrument: Reichert - Jung KF80 |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Temperature | Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 100,000 times magnification |
| Date | Sep 24, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 32 / Average electron dose: 15 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 48327 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan 626 / Specimen holder model: GATAN LIQUID NITROGEN |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)