[English] 日本語
 Yorodumi
Yorodumi- EMDB-53153: Pseudomonas aeruginosa polynucleotide phosphorylase in complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas aeruginosa polynucleotide phosphorylase in complex with recognition site of RNase E | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polynucleotide phosphorylase / ribonuclease E / RNA degradosome / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonuclease E / ribonuclease E activity / polyribonucleotide nucleotidyltransferase / polyribonucleotide nucleotidyltransferase activity / RNA catabolic process / tRNA processing / mRNA catabolic process / RNA nuclease activity / RNA processing / cytoplasmic side of plasma membrane ...ribonuclease E / ribonuclease E activity / polyribonucleotide nucleotidyltransferase / polyribonucleotide nucleotidyltransferase activity / RNA catabolic process / tRNA processing / mRNA catabolic process / RNA nuclease activity / RNA processing / cytoplasmic side of plasma membrane / rRNA processing / 3'-5'-RNA exonuclease activity / tRNA binding / rRNA binding / magnesium ion binding / RNA binding / zinc ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Paris G / Luisi BF | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2025 Journal: Nucleic Acids Res / Year: 2025Title: A multi-dentate, cooperative interaction between endo- and exo-ribonucleases within the bacterial RNA degradosome. Authors: Giulia Paris / Kai Katsuya-Gaviria / Hannah Clarke / Margaret Johncock / Tom Dendooven / Aleksei Lulla / Ben F Luisi /  Abstract: In Escherichia coli and numerous other bacteria, two of the principal enzymes mediating messenger RNA decay and RNA processing-RNase E, an endoribonuclease, and polynucleotide phosphorylase (PNPase), ...In Escherichia coli and numerous other bacteria, two of the principal enzymes mediating messenger RNA decay and RNA processing-RNase E, an endoribonuclease, and polynucleotide phosphorylase (PNPase), an exoribonuclease-assemble into a multi-enzyme complex known as the RNA degradosome. While RNase E forms a homotetramer and PNPase a homotrimer, it remains unclear how these two enzymes interact within the RNA degradosome to potentially satisfy all mutual recognition sites. In this study, we used cryo-EM, biochemistry, and biophysical studies to discover and characterize a new binding mode for PNPase encompassing two or more motifs that are necessary and sufficient for strong interaction with RNase E. While a similar interaction is seen in Salmonella enterica, a different recognition mode arose for Pseudomonas aeruginosa, illustrating the evolutionary drive to maintain physical association of the two ribonucleases. The data presented here suggest a model for the quaternary organization of the RNA degradosome of E. coli, where one PNPase trimer interacts with one RNase E protomer. Conformational transitions are predicted to facilitate substrate capture and transfer to catalytic centres. The model suggests how the endo- and exo-ribonucleases might cooperate in cellular RNA turnover and recruitment of regulatory RNA by the degradosome assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_53153.map.gz emd_53153.map.gz | 89.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-53153-v30.xml emd-53153-v30.xml emd-53153.xml emd-53153.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_53153_fsc.xml emd_53153_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_53153.png emd_53153.png | 73.2 KB | ||
| Filedesc metadata |  emd-53153.cif.gz emd-53153.cif.gz | 7.1 KB | ||
| Others |  emd_53153_half_map_1.map.gz emd_53153_half_map_1.map.gz emd_53153_half_map_2.map.gz emd_53153_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-53153 http://ftp.pdbj.org/pub/emdb/structures/EMD-53153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53153 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-53153 | HTTPS FTP |
-Related structure data
| Related structure data |  9qh3MC  9qh0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_53153.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_53153.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: One of the two half-maps
| File | emd_53153_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the two half-maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second of the two half-maps
| File | emd_53153_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second of the two half-maps | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Polynucleotide phosphorylase in complex with recognition site fro...
| Entire | Name: Polynucleotide phosphorylase in complex with recognition site from ribonuclease E |
|---|---|
| Components |
|
-Supramolecule #1: Polynucleotide phosphorylase in complex with recognition site fro...
| Supramolecule | Name: Polynucleotide phosphorylase in complex with recognition site from ribonuclease E type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Complex prepared by co-expression and chromatographic purification |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Polyribonucleotide nucleotidyltransferase
| Macromolecule | Name: Polyribonucleotide nucleotidyltransferase / type: protein_or_peptide / ID: 1 / Details: catalytic core, without S1 and KH domains / Number of copies: 3 / Enantiomer: LEVO / EC number: polyribonucleotide nucleotidyltransferase |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 59.727867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNPVTKQFQF GQSTVTLETG RIARQATGAV LVTMDDVSVL VTVVGAKSPA EGRDFFPLSV HYQEKTYAAG RIPGGFFKRE GRPSEKETL TSRLIDRPIR PLFPEGFMNE VQVVCTVVST NKKSDPDIAA MIGTSAALAI SGIPFAGPIG AARVGFHPEI G YILNPTYE ...String: MNPVTKQFQF GQSTVTLETG RIARQATGAV LVTMDDVSVL VTVVGAKSPA EGRDFFPLSV HYQEKTYAAG RIPGGFFKRE GRPSEKETL TSRLIDRPIR PLFPEGFMNE VQVVCTVVST NKKSDPDIAA MIGTSAALAI SGIPFAGPIG AARVGFHPEI G YILNPTYE QLQSSSLDMV VAGTEDAVLM VESEADELTE DQMLGAVLFA HDEFQAVIRA VKELAAEAGK PAWDWKAPAE NT VLVNAIK AELGEAISQA YTITIKQDRY NRLGELRDQA VALFAGEEEG KFPASEVKDV FGLLEYRTVR ENIVNGKPRI DGR DTRTVR PLRIEVGVLG KTHGSALFTR GETQALVVAT LGTARDAQLL DTLEGERKDA FMLHYNFPPF SVGECGRMGS PGRR EIGHG RLARRGVAAM LPTQDEFPYT IRVVSEITES NGSSSMASVC GASLALMDAG VPVKAPVAGI AMGLVKEGEK FAVLT DILG DEDHLGDMDF KVAGTDKGVT ALQMDIKING ITEEIMEIAL GQALEARLNI LGQMNQVIAK PRAELSENAP UniProtKB: Polyribonucleotide nucleotidyltransferase |
-Macromolecule #2: Ribonuclease E
| Macromolecule | Name: Ribonuclease E / type: protein_or_peptide / ID: 2 / Details: PNPase recognition site from RNase E / Number of copies: 1 / Enantiomer: LEVO / EC number: ribonuclease E |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PAO1 (bacteria) Pseudomonas aeruginosa PAO1 (bacteria) |
| Molecular weight | Theoretical: 2.911264 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VPANATGRAL NDPREKRRLQ REAER UniProtKB: Ribonuclease E |
-Macromolecule #3: ADENOSINE-5'-MONOPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-MONOPHOSPHATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: A |
|---|---|
| Molecular weight | Theoretical: 347.221 Da |
| Chemical component information | 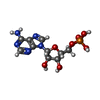 ChemComp-A: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 45 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.6 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris-HCl pH 8.0, 25 mM MgCl2, 150 mM KCl, 1 mM TCEP |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.2 K / Instrument: FEI VITROBOT MARK IV / Details: blotting force -4, 3 sec blot time. |
| Details | Purified PNPase core and RNE-muGFP-CHis proteins were mixed in 1:2 ratio and their complex were separated on the Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated with Cryo-EM buffer (20 mM Tris-HCl pH 8.0, 25 mM MgCl2, 150 mM KCl, 1 mM TCEP). Peak fractions were combined, the protein was concentrated to 15 microM using Amicon Ultra concentrator with 10 kDa cut-off (Millipore) and used to prepare Cryo-EM grids. The samples were mixed with CHAPSO (3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate) at a final concentration of 8 mM |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 4000 / Average exposure time: 4.39 sec. / Average electron dose: 53.94 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | phenix refine and manual rebuilding using COOT |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-9qh3: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































