[English] 日本語
 Yorodumi
Yorodumi- EMDB-5252: electron cryo-tomography reconstruction and subvolume averaging o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5252 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | electron cryo-tomography reconstruction and subvolume averaging of the basal body triplet from Chlamydomonas reinhardtii | |||||||||
 Map data Map data | averaged microtubule triplet from basal body from Chlamydomonas reinhardtii | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | basal body / centriole / electron cryo-tomography / three-dimensional reconstruction | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 33.0 Å | |||||||||
 Authors Authors | Li S / Fernandez JJ / Marshall WF / Agard DA | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2012 Journal: EMBO J / Year: 2012Title: Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. Authors: Sam Li / Jose-Jesus Fernandez / Wallace F Marshall / David A Agard /  Abstract: Basal bodies and centrioles play central roles in microtubule (MT)-organizing centres within many eukaryotes. They share a barrel-shaped cylindrical structure composed of nine MT triplet blades. ...Basal bodies and centrioles play central roles in microtubule (MT)-organizing centres within many eukaryotes. They share a barrel-shaped cylindrical structure composed of nine MT triplet blades. Here, we report the structure of the basal body triplet at 33 Å resolution obtained by electron cryo-tomography and 3D subtomogram averaging. By fitting the atomic structure of tubulin into the EM density, we built a pseudo-atomic model of the tubulin protofilaments at the core of the triplet. The 3D density map reveals additional densities that represent non-tubulin proteins attached to the triplet, including a large inner circular structure in the basal body lumen, which functions as a scaffold to stabilize the entire basal body barrel. We found clear longitudinal structural variations along the basal body, suggesting a sequential and coordinated assembly mechanism. We propose a model in which δ-tubulin and other components participate in the assembly of the basal body. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5252.map.gz emd_5252.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5252-v30.xml emd-5252-v30.xml emd-5252.xml emd-5252.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5252_1.tif emd_5252_1.tif | 235.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5252 http://ftp.pdbj.org/pub/emdb/structures/EMD-5252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5252 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5252 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5252.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5252.map.gz / Format: CCP4 / Size: 6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | averaged microtubule triplet from basal body from Chlamydomonas reinhardtii | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
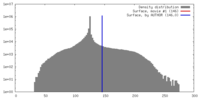
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : basal body microtubule triplet
| Entire | Name: basal body microtubule triplet |
|---|---|
| Components |
|
-Supramolecule #1000: basal body microtubule triplet
| Supramolecule | Name: basal body microtubule triplet / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 15.1 MDa Method: estimation based on the volume assuming the protein density is 1.41g per cm3 |
-Supramolecule #1: basal body
| Supramolecule | Name: basal body / type: organelle_or_cellular_component / ID: 1 / Name.synonym: basal body / Number of copies: 2 / Oligomeric state: singlet / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Buffer | pH: 8 / Details: 10mM Tris 1mM EDTA |
|---|---|
| Grid | Details: 300 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 93 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 91 K / Max: 93 K / Average: 93 K |
| Specialist optics | Energy filter - Name: Gatan GIF2000 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 25.0 eV |
| Details | defocus maximum value is 24000nm |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Average electron dose: 80 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 46153 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus min: 9.0 µm / Nominal magnification: 34000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The tomographic tilt series were taken by UCSF Tomography program. Average number of tilts used in the 3D reconstructions: 82. Average tomographic tilt angle increment: 2. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 33.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Priism / Details: Final map was average of 1644 subvolumes |
| CTF correction | Details: each image, Wiener filter |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | Protocol: rigid body. The atomic structure was first manually fitting into EM density by program O and the fitting was refined by Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation coefficiet |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)