[English] 日本語
 Yorodumi
Yorodumi- EMDB-52200: Extracellular components BamHIJK of the Bacteroides thetaiotaomic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Extracellular components BamHIJK of the Bacteroides thetaiotaomicron BAM machinery | |||||||||
 Map data Map data | unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lipoproteins / Outer membrane protein biogenesis / Beta-barrel assembly machinery / BAM / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Silale A / van den Berg B | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2025 Journal: Nat Microbiol / Year: 2025Title: Structure of a distinct β-barrel assembly machinery complex in the Bacteroidota. Authors: Augustinas Silale / Mariusz Madej / Katarzyna Mikruta / Andrew M Frey / Adam J Hart / Arnaud Baslé / Carsten Scavenius / Jan J Enghild / Matthias Trost / Robert P Hirt / Bert van den Berg /    Abstract: The Gram-negative β-barrel assembly machinery (BAM) complex catalyses the folding and membrane insertion of newly synthesized β-barrel outer membrane proteins. The BAM is structurally conserved, ...The Gram-negative β-barrel assembly machinery (BAM) complex catalyses the folding and membrane insertion of newly synthesized β-barrel outer membrane proteins. The BAM is structurally conserved, but most studies have focused on Gammaproteobacteria. Here, using single-particle cryogenic electron microscopy, quantitative proteomics and functional assays, we show that the BAM complex is distinct within the Bacteroidota. Cryogenic electron microscopy structures of BAM complexes from the human gut symbiont Bacteroides thetaiotaomicron (3.3 Å) and the human oral pathogen Porphyromonas gingivalis (3.2 Å) show similar, seven-component complexes of ~325 kDa. The complexes are mostly extracellular and comprise canonical BamA and BamD; an integral, essential outer membrane protein, BamG, that associates with BamA; and four surface-exposed lipoproteins: BamH-K. Absent from the BAM in Pseudomonadota, BamG-K form a large, extracellular dome that may confer additional functionality to enable the folding and assembly of β-barrel-surface-exposed lipoprotein complexes that are a hallmark of the Bacteroidota. Our findings develop our understanding of fundamental biological processes in an important bacterial phylum. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_52200.map.gz emd_52200.map.gz | 170.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-52200-v30.xml emd-52200-v30.xml emd-52200.xml emd-52200.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_52200_fsc.xml emd_52200_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_52200.png emd_52200.png | 90.7 KB | ||
| Masks |  emd_52200_msk_1.map emd_52200_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-52200.cif.gz emd-52200.cif.gz | 7.8 KB | ||
| Others |  emd_52200_additional_1.map.gz emd_52200_additional_1.map.gz emd_52200_half_map_1.map.gz emd_52200_half_map_1.map.gz emd_52200_half_map_2.map.gz emd_52200_half_map_2.map.gz | 324 MB 317.9 MB 317.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-52200 http://ftp.pdbj.org/pub/emdb/structures/EMD-52200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-52200 | HTTPS FTP |
-Related structure data
| Related structure data |  9hisMC  9hivC  9hj3C  9hjmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_52200.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_52200.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_52200_msk_1.map emd_52200_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharpened map
| File | emd_52200_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_52200_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_52200_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Extracellular components BamGHIJ of the Bacteroides thetaiotaomic...
| Entire | Name: Extracellular components BamGHIJ of the Bacteroides thetaiotaomicron BAM complex |
|---|---|
| Components |
|
-Supramolecule #1: Extracellular components BamGHIJ of the Bacteroides thetaiotaomic...
| Supramolecule | Name: Extracellular components BamGHIJ of the Bacteroides thetaiotaomicron BAM complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #4, #1-#3 |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 153 KDa |
-Macromolecule #1: DUF6242 domain-containing protein
| Macromolecule | Name: DUF6242 domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 55.108051 KDa |
| Sequence | String: MRIKFLSIIA SFFMVSFVIT SCLDNDNEVN YSPDATIRAF ELDTIGYGVN YKFTIDQVSR LIYNVDSLPV NADTIINSIL IKTLTTASG IVTMKDQNDQ DSIVNINDSI DLTKYVNATE KNNFLVLKVW APNMEVQNEY KVNIRMHTMV PDSLSWGKDP I ANNPVRNT ...String: MRIKFLSIIA SFFMVSFVIT SCLDNDNEVN YSPDATIRAF ELDTIGYGVN YKFTIDQVSR LIYNVDSLPV NADTIINSIL IKTLTTASG IVTMKDQNDQ DSIVNINDSI DLTKYVNATE KNNFLVLKVW APNMEVQNEY KVNIRMHTMV PDSLSWGKDP I ANNPVRNT AEKQKVVTLG DKILLFAQNN EIYSTAIPAG SPTDRLNYGQ KWDKETTGKL PDGADVTSII RFVDKLYLLT KN KEVYNSN DGLTWTKDEV LNSDGVSVTN LITSFSDSDG SNHKKINGIA GIVEINGEKY FSFAEKDVTW EKDIDKLTVV PAE FPINNL SADVYATESG TLNAIVVGNT EDGLDNDTAT VVWASEDGKA WIPMEIPSNN NCPKLVDPSI IHYNDAFYIC GKET KDDAK GFQKFYTSPT LLVWKGVDRM FMLPGILPPV KLEGGVTQHP SLHESSFKGK EVNYTMVVDR NHYIWMVGGQ GIDKI WRGR VNKLGFLIQ UniProtKB: Exo-alpha-sialidase |
-Macromolecule #2: Peptidyl-prolyl cis-trans isomerase
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 21.884557 KDa |
| Sequence | String: MSKKIYLFSL VLLALAFVSC SETEEVGKYD NWRARNEAFI DSLANVYATA SGRGGLERIE MLTAPGNYIY YKEMEPMTDH VVKAGNPKY TDYVKVYYKG TNILGEYFDG NFKGDNPVVD GKDPSEGDSP TTIFQVSGVI TGWGEVLQRM EVGDRWKVYI P WDYAYGSS ...String: MSKKIYLFSL VLLALAFVSC SETEEVGKYD NWRARNEAFI DSLANVYATA SGRGGLERIE MLTAPGNYIY YKEMEPMTDH VVKAGNPKY TDYVKVYYKG TNILGEYFDG NFKGDNPVVD GKDPSEGDSP TTIFQVSGVI TGWGEVLQRM EVGDRWKVYI P WDYAYGSS GTTGILGYSA LVFDITLLDF ANTEAELK UniProtKB: Peptidyl-prolyl cis-trans isomerase |
-Macromolecule #3: DUF4827 domain-containing protein
| Macromolecule | Name: DUF4827 domain-containing protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 24.712709 KDa |
| Sequence | String: MKKLVFLFLS LLAAGGIFQA CDDSKTYAEM LEDEKNAVNK FIKDKGIRII SQDEFEKNDT VTNLERNEYV ALSDGVYMQI VDRGSAENK TDTFANNNEI CVRYIEEDIM TRDTTCFNVF LEEWGDANQL YTNPAVFRYV AEGSYVYGTF IQMDYYWASY Y QSTAVPAG ...String: MKKLVFLFLS LLAAGGIFQA CDDSKTYAEM LEDEKNAVNK FIKDKGIRII SQDEFEKNDT VTNLERNEYV ALSDGVYMQI VDRGSAENK TDTFANNNEI CVRYIEEDIM TRDTTCFNVF LEEWGDANQL YTNPAVFRYV AEGSYVYGTF IQMDYYWASY Y QSTAVPAG WLLALPFVRN YAHVRLIVPS KVGHSSAQQY VNPYYYDIWT FSKALN UniProtKB: DUF4827 domain-containing protein |
-Macromolecule #4: DUF4270 domain-containing protein
| Macromolecule | Name: DUF4270 domain-containing protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 58.264816 KDa |
| Sequence | String: CDDNTGGLGL GMFPGNDQNI KGKLSTFDVT TESVKTGDIY AKTNIGYIGK FTDETFGTYQ AGFLAQLNCP DGLTFPEPYK EVTDASGNV ISATGRMVVD DKDPENKDVT FIKDGNQIIG NIRAVELYLW YDSYFGDSLT ACRLSVYELG GNGKETLNLD N AYYTDINP ...String: CDDNTGGLGL GMFPGNDQNI KGKLSTFDVT TESVKTGDIY AKTNIGYIGK FTDETFGTYQ AGFLAQLNCP DGLTFPEPYK EVTDASGNV ISATGRMVVD DKDPENKDVT FIKDGNQIIG NIRAVELYLW YDSYFGDSLT ACRLSVYELG GNGKETLNLD N AYYTDINP EDFYDSQNIL GTKAYTAVDL SVKDSIRNLS TYVPSVHIAF KEDIATRVGG NILTAARKAK NADKEFNSQL FR EAFQGIY VKSDYGDGTV LYIDQPQMNV VYKCYATDSI TGKKLQKKDG SGKDSTYYSY RVFATTREVI QANQLKNDPE RID ALIKED KNTYLKSPAG IFTEATLPIS DIQNELTGDT LNAVKLTFTN YNQTGDKKFG MAIPSTVMLV RKKFQDSFFK DNKL SDGVS SYLTSHTSST NQYVFSNITK LVNACIAEKE EAKKNAGSSW DETKWLQENP DWNKVVLIPV LVTYDSSNTT TGQAN IIRI QHDLKPGYVR LKGGSLGKTN PDYKLKLEVI STDFGLTTKS N UniProtKB: DUF4270 domain-containing protein |
-Macromolecule #5: N-TRIDECANOIC ACID
| Macromolecule | Name: N-TRIDECANOIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: TDA |
|---|---|
| Molecular weight | Theoretical: 214.344 Da |
| Chemical component information | 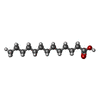 ChemComp-TDA: |
-Macromolecule #6: (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate
| Macromolecule | Name: (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate / type: ligand / ID: 6 / Number of copies: 1 / Formula: Z41 |
|---|---|
| Molecular weight | Theoretical: 568.911 Da |
| Chemical component information |  ChemComp-Z41: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 13558 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Details | AlphaFold2 models were docked using Phenix. | ||||||||||
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 58 | ||||||||||
| Output model |  PDB-9his: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)

























































