+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5215 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Anthrax toxin PA63 in complex with 1G3 | |||||||||
 Map data Map data | Anthrax toxin component PA63 in complex with neutralizing antibody 1G3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | anthrax toxin protective antigen neutralizing antibody | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 33.0 Å | |||||||||
 Authors Authors | Radjainia M / Hyun J / Leysath CE / Leppla SH / Mitra AK | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2010 Journal: Proc Natl Acad Sci U S A / Year: 2010Title: Anthrax toxin-neutralizing antibody reconfigures the protective antigen heptamer into a supercomplex. Authors: Mazdak Radjainia / Jae-Kyung Hyun / Clinton E Leysath / Stephen H Leppla / Alok K Mitra /  Abstract: The tripartite protein exotoxin secreted by Bacillus anthracis, a major contributor to its virulence and anthrax pathogenesis, consists of binary complexes of the protective antigen (PA) heptamer ...The tripartite protein exotoxin secreted by Bacillus anthracis, a major contributor to its virulence and anthrax pathogenesis, consists of binary complexes of the protective antigen (PA) heptamer (PA63h), produced by proteolytic cleavage of PA, together with either lethal factor or edema factor. The mouse monoclonal anti-PA antibody 1G3 was previously shown to be a potent antidote that shares F(C) domain dependency with the human monoclonal antibody MDX-1303 currently under clinical development. Here we demonstrate that 1G3 instigates severe perturbation of the PA63h structure and creates a PA supercomplex as visualized by electron microscopy. This phenotype, produced by the unconventional mode of antibody action, highlights the feasibility for optimization of vaccines based on analogous structural modification of PA63h as an additional strategy for future remedies against anthrax. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5215.map.gz emd_5215.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5215-v30.xml emd-5215-v30.xml emd-5215.xml emd-5215.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5215_1.png emd_5215_1.png | 89.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5215 http://ftp.pdbj.org/pub/emdb/structures/EMD-5215 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5215 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5215 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5215.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5215.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anthrax toxin component PA63 in complex with neutralizing antibody 1G3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Anthrax toxin component PA63 in complex with 1G3
| Entire | Name: Anthrax toxin component PA63 in complex with 1G3 |
|---|---|
| Components |
|
-Supramolecule #1000: Anthrax toxin component PA63 in complex with 1G3
| Supramolecule | Name: Anthrax toxin component PA63 in complex with 1G3 / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 1.1 MDa / Method: Size-exclusion chromatography |
-Macromolecule #1: toxin
| Macromolecule | Name: toxin / type: protein_or_peptide / ID: 1 / Name.synonym: protective antigen / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 9 / Details: 150mM NaCl, 50mM CHES |
| Staining | Type: NEGATIVE Details: Holey grids with samples negatively stained across holes with 1% uranyl acetate |
| Grid | Details: Quantifoil R1.2/1.3 |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 97000 times magnification |
| Date | Sep 1, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 13 µm / Number real images: 40 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder: single-tilt / Specimen holder model: OTHER / Tilt angle max: 45 |
- Image processing
Image processing
| Details | The particles were manually selected. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 33.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: XMIPP / Number images used: 3459 |
| Final two d classification | Number classes: 20 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera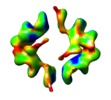




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















