[English] 日本語
 Yorodumi
Yorodumi- EMDB-51478: Cryo-EM structure of Sporosarcina pasteurii urease inhibited by NBPTO -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Sporosarcina pasteurii urease inhibited by NBPTO | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | urease / urea / nickel / NBPTO / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationurease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Mazzei L / Tria G / Ciurli S / Cianci M | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Int J Biol Macromol / Year: 2024 Journal: Int J Biol Macromol / Year: 2024Title: Exploring the conformational space of the mobile flap in Sporosarcina pasteurii urease by cryo-electron microscopy. Authors: Luca Mazzei / Giancarlo Tria / Stefano Ciurli / Michele Cianci /  Abstract: To fully understand enzymatic dynamics, it is essential to explore the complete conformational space of a biological catalyst. The catalytic mechanism of the nickel-dependent urease, the most ...To fully understand enzymatic dynamics, it is essential to explore the complete conformational space of a biological catalyst. The catalytic mechanism of the nickel-dependent urease, the most efficient enzyme known, holds significant relevance for medical, pharmaceutical, and agro-environmental applications. A critical aspect of urease function is the conformational change of a helix-turn-helix motif that covers the active site cavity, known as the mobile flap. This motif has been observed in either an open or a closed conformation through X-ray crystallography studies and has been proposed to stabilize the coordination of a urea molecule to the essential dinuclear Ni(II) cluster in the active site, a requisite for subsequent substrate hydrolysis. This study employs cryo-electron microscopy (cryo-EM) to investigate the transient states within the conformational space of the mobile flap, devoid of the possible constraints of crystallization conditions and solid-state effects. By comparing two cryo-EM structures of Sporosarcina pasteurii urease, one in its native form and the other inhibited by N-(n-butyl) phosphoric triamide (NBPTO), we have unprecedently identified an intermediate state between the open and the catalytically efficient closed conformation of the helix-turn-helix motif, suggesting a role of its tip region in this transition between the two states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51478.map.gz emd_51478.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51478-v30.xml emd-51478-v30.xml emd-51478.xml emd-51478.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51478_fsc.xml emd_51478_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_51478.png emd_51478.png | 76.6 KB | ||
| Filedesc metadata |  emd-51478.cif.gz emd-51478.cif.gz | 6.1 KB | ||
| Others |  emd_51478_additional_1.map.gz emd_51478_additional_1.map.gz emd_51478_half_map_1.map.gz emd_51478_half_map_1.map.gz emd_51478_half_map_2.map.gz emd_51478_half_map_2.map.gz | 897.2 KB 59 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51478 http://ftp.pdbj.org/pub/emdb/structures/EMD-51478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51478 | HTTPS FTP |
-Related structure data
| Related structure data |  9gnrMC  9gmlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_51478.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51478.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_51478_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_51478_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_51478_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : urease
| Entire | Name: urease |
|---|---|
| Components |
|
-Supramolecule #1: urease
| Supramolecule | Name: urease / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Urease subunit gamma
| Macromolecule | Name: Urease subunit gamma / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) |
| Molecular weight | Theoretical: 11.134895 KDa |
| Sequence | String: (CXM)HLNPAEKEK LQIFLASELA LKRKARGLKL NYPEAVAIIT SFIMEGARDG KTVAMLMEEG KHVLTRDDVM EGVPEM IDD IQAEATFPDG TKLVTVHNPI S UniProtKB: Urease subunit gamma |
-Macromolecule #2: Urease subunit beta
| Macromolecule | Name: Urease subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) |
| Molecular weight | Theoretical: 13.529061 KDa |
| Sequence | String: NYIVPGEYRV AEGEIEINAG REKTTIRVSN TGDRPIQVGS HIHFVEVNKE LLFDRAEGIG RRLNIPSGTA ARFEPGEEME VELTELGGN REVFGISDLT NGSVDNKELI LQRAKELGYK GVE UniProtKB: Urease subunit beta |
-Macromolecule #3: Urease subunit alpha
| Macromolecule | Name: Urease subunit alpha / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  Sporosarcina pasteurii (bacteria) Sporosarcina pasteurii (bacteria) |
| Molecular weight | Theoretical: 61.575648 KDa |
| Sequence | String: MKINRQQYAE SYGPTVGDQV RLADTDLWIE VEKDYTTYGD EANFGGGKVL REGMGENGTY TRTENVLDLL LTNALILDYT GIYKADIGV KDGYIVGIGK GGNPDIMDGV TPNMIVGTAT EVIAAEGKIV TAGGIDTHVH FINPDQVDVA LANGITTLFG G GTGPAEGS ...String: MKINRQQYAE SYGPTVGDQV RLADTDLWIE VEKDYTTYGD EANFGGGKVL REGMGENGTY TRTENVLDLL LTNALILDYT GIYKADIGV KDGYIVGIGK GGNPDIMDGV TPNMIVGTAT EVIAAEGKIV TAGGIDTHVH FINPDQVDVA LANGITTLFG G GTGPAEGS KATTVTPGPW NIEKMLKSTE GLPINVGILG KGHGSSIAPI MEQIDAGAAG L(KCX)IHEDWGAT PASIDRSL T VADEADVQVA IHSDTLNEAG FLEDTLRAIN GRVIHSFHVE GAGGGHAPDI MAMAGHPNVL PSSTNPTRPF TVNTIDEHL DMLMVCHHLK QNIPEDVAFA DSRIRPETIA AEDILHDLGI ISMMSTDALA MGRAGEMVLR TWQTADKMKK QRGPLAEEKN GSDNFRAKR YVSKYTINPA IAQGIAHEVG SIEEGKFADL VLWEPKFFGV KADRVIKGGI IAYAQIGDPS ASIPTPQPVM G RRMYGTVG DLIHDTNITF MSKSSIQQGV PAKLGLKRRI GTVKNCRNIG KKDMKWNDVT TDIDINPETY EVKVDGEVLT CE PVKELPM AQRYFLF UniProtKB: Urease subunit alpha |
-Macromolecule #4: NICKEL (II) ION
| Macromolecule | Name: NICKEL (II) ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: NI |
|---|---|
| Molecular weight | Theoretical: 58.693 Da |
| Chemical component information |  ChemComp-NI: |
-Macromolecule #5: DIAMIDOPHOSPHATE
| Macromolecule | Name: DIAMIDOPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: 2PA |
|---|---|
| Molecular weight | Theoretical: 96.026 Da |
| Chemical component information | 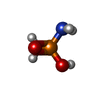 ChemComp-2PA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 50 mM HEPES buffer, at pH 7.5, also containing 5 mM NBPTO |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 1459548 / Average exposure time: 3.2 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-9gnr: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































