[English] 日本語
 Yorodumi
Yorodumi- EMDB-5130: 3D cryo-EM Structure of Archaeal 20S Proteasome in Complex with t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5130 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D cryo-EM Structure of Archaeal 20S Proteasome in Complex with the C-terminus of PAN | |||||||||
 Map data Map data | This is the 3D volume of T.acidophilum 20S proteasome in complex with a hybrid activator of PA26 and PAN | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Proteasome / proteasomal ATPase / protein degradation / AAA ATPase / x-ray crystallography / electron cryomicroscopy | |||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Yu Y / Smith DM / Kim H / Rodriguez V / Goldberg AL / Cheng Y | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2010 Journal: EMBO J / Year: 2010Title: Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. Authors: Yadong Yu / David M Smith / Ho Min Kim / Victor Rodriguez / Alfred L Goldberg / Yifan Cheng /  Abstract: Protein degradation in the 20S proteasome is regulated in eukaryotes by the 19S ATPase complex and in archaea by the homologous PAN ATPase ring complex. Subunits of these hexameric ATPases contain on ...Protein degradation in the 20S proteasome is regulated in eukaryotes by the 19S ATPase complex and in archaea by the homologous PAN ATPase ring complex. Subunits of these hexameric ATPases contain on their C-termini a conserved hydrophobic-tyrosine-X (HbYX) motif that docks into pockets in the 20S to stimulate the opening of a gated substrate entry channel. Here, we report the crystal structure of the archaeal 20S proteasome in complex with the C-terminus of the archaeal proteasome regulatory ATPase, PAN. This structure defines the detailed interactions between the critical C-terminal HbYX motif and the 20S alpha-subunits and indicates that the intersubunit pocket in the 20S undergoes an induced-fit conformational change on binding of the HbYX motif. This structure together with related mutagenesis data suggest how in eukaryotes certain proteasomal ATPases bind to specific pockets in an asymmetrical manner to regulate gate opening. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5130.map.gz emd_5130.map.gz | 9.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5130-v30.xml emd-5130-v30.xml emd-5130.xml emd-5130.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5130_1.tif emd_5130_1.tif | 1.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5130 http://ftp.pdbj.org/pub/emdb/structures/EMD-5130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5130 | HTTPS FTP |
-Validation report
| Summary document |  emd_5130_validation.pdf.gz emd_5130_validation.pdf.gz | 78.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5130_full_validation.pdf.gz emd_5130_full_validation.pdf.gz | 77.9 KB | Display | |
| Data in XML |  emd_5130_validation.xml.gz emd_5130_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5130 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5130 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5130 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5130 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5130.map.gz / Format: CCP4 / Size: 9.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5130.map.gz / Format: CCP4 / Size: 9.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D volume of T.acidophilum 20S proteasome in complex with a hybrid activator of PA26 and PAN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 1.79103 Å / Y: 1.79103 Å / Z: 1.79102 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
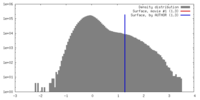
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Archaeal 20S proteasome in complex with a hybrid of PA26 and PAN
| Entire | Name: Archaeal 20S proteasome in complex with a hybrid of PA26 and PAN |
|---|---|
| Components |
|
-Supramolecule #1000: Archaeal 20S proteasome in complex with a hybrid of PA26 and PAN
| Supramolecule | Name: Archaeal 20S proteasome in complex with a hybrid of PA26 and PAN type: sample / ID: 1000 / Details: Proteins were expressed from E.coli. Oligomeric state: 20S proteasome is composed of 4 heptamer rings coaxially stacked to which heptamers of the PA26 PAN hybrid cap on both ends Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 1.1 MDa |
-Macromolecule #1: 20S proteasome, PA26-PAN
| Macromolecule | Name: 20S proteasome, PA26-PAN / type: protein_or_peptide / ID: 1 / Name.synonym: 20S proteasome, PA26-PAN / Number of copies: 1 Oligomeric state: 2 heptamers of 20S alpha subunit, 2 heptamers of 20S beta subunit, and 2 heptamers of PA26-PAN Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Molecular weight | Theoretical: 1.1 MDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8.5 Details: 50 mM Tris pH 8.5, 1mM DTT, 10 mM MgCl2, 5% Glycerol |
| Grid | Details: 400 mesh Cu grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: blot for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 90 K / Max: 90 K / Average: 90 K |
| Details | data was collected using Leginon |
| Date | Sep 17, 2007 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN (4k x 4k) / Average electron dose: 20 e/Å2 / Details: images were recorded on CCD camera |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder: Gatan CT-3500 / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were manually selected |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: FREALIGN / Details: D7 symmetry was applied / Number images used: 13020 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)