[English] 日本語
 Yorodumi
Yorodumi- EMDB-50804: Density for a protein trimer unit bound to RNA from a single-laye... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Density for a protein trimer unit bound to RNA from a single-layered nucleoprotein-RNA helical assembly from Marburg virus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-binding protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral RNA genome packaging / helical viral capsid / viral budding via host ESCRT complex / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / RNA binding Similarity search - Function | |||||||||
| Biological species |  Marburg virus - Musoke, Kenya, 1980 / Marburg virus - Musoke, Kenya, 1980 /  Marburg virus - Musoke Marburg virus - Musoke | |||||||||
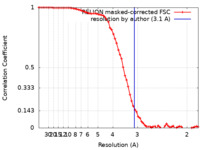
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zinzula L / Beck F / Camasta M / Bohn S / Liu C / Morado D / Bracher A / Plitzko JM / Baumeister W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structure of single-layered nucleoprotein-RNA complex from Marburg virus. Authors: Luca Zinzula / Florian Beck / Marianna Camasta / Stefan Bohn / Chuan Liu / Dustin Morado / Andreas Bracher / Juergen M Plitzko / Wolfgang Baumeister /    Abstract: Marburg virus (MARV) causes lethal hemorrhagic fever in humans, posing a threat to global health. We determined by cryogenic electron microscopy (cryo-EM) the MARV helical ribonucleoprotein (RNP) ...Marburg virus (MARV) causes lethal hemorrhagic fever in humans, posing a threat to global health. We determined by cryogenic electron microscopy (cryo-EM) the MARV helical ribonucleoprotein (RNP) complex structure in single-layered conformation, which differs from the previously reported structure of a double-layered helix. Our findings illuminate novel RNP interactions and expand knowledge on MARV genome packaging and nucleocapsid assembly, both processes representing attractive targets for the development of antiviral therapeutics against MARV disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50804.map.gz emd_50804.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50804-v30.xml emd-50804-v30.xml emd-50804.xml emd-50804.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50804_fsc.xml emd_50804_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_50804.png emd_50804.png | 71.5 KB | ||
| Masks |  emd_50804_msk_1.map emd_50804_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50804.cif.gz emd-50804.cif.gz | 5.5 KB | ||
| Others |  emd_50804_half_map_1.map.gz emd_50804_half_map_1.map.gz emd_50804_half_map_2.map.gz emd_50804_half_map_2.map.gz | 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50804 http://ftp.pdbj.org/pub/emdb/structures/EMD-50804 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50804 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50804 | HTTPS FTP |
-Validation report
| Summary document |  emd_50804_validation.pdf.gz emd_50804_validation.pdf.gz | 890.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50804_full_validation.pdf.gz emd_50804_full_validation.pdf.gz | 890.2 KB | Display | |
| Data in XML |  emd_50804_validation.xml.gz emd_50804_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_50804_validation.cif.gz emd_50804_validation.cif.gz | 17 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50804 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50804 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50804 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50804 | HTTPS FTP |
-Related structure data
| Related structure data |  9fvdC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50804.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50804.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
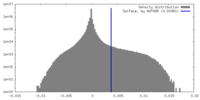
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50804_msk_1.map emd_50804_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_50804_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_50804_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical assembly of nucleoprotein-RNA complex from Marburg virus
| Entire | Name: Helical assembly of nucleoprotein-RNA complex from Marburg virus |
|---|---|
| Components |
|
-Supramolecule #1: Helical assembly of nucleoprotein-RNA complex from Marburg virus
| Supramolecule | Name: Helical assembly of nucleoprotein-RNA complex from Marburg virus type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Marburg virus - Musoke, Kenya, 1980 Marburg virus - Musoke, Kenya, 1980 |
-Macromolecule #1: N protein from Marburg virus
| Macromolecule | Name: N protein from Marburg virus / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Marburg virus - Musoke Marburg virus - Musoke |
| Recombinant expression | Organism:  |
| Sequence | String: GMDLHSLLEL GTKPTAPHVR NKKVILFDTN HQVSICNQII DAINSGIDLG DLLEGGLLTL CVEHYYNSDK DKFNTSPIAK YLRDAGYEFD VIKNADATRF LDVIPNEPHY SPLILALKTL ESTESQRGRI GLFLSFCSLF LPKLVVGDRA SIEKALRQVT VHQEQGIVTY ...String: GMDLHSLLEL GTKPTAPHVR NKKVILFDTN HQVSICNQII DAINSGIDLG DLLEGGLLTL CVEHYYNSDK DKFNTSPIAK YLRDAGYEFD VIKNADATRF LDVIPNEPHY SPLILALKTL ESTESQRGRI GLFLSFCSLF LPKLVVGDRA SIEKALRQVT VHQEQGIVTY PNHWLTTGHM KVIFGILRSS FILKFVLIHQ GVNLVTGHDA YDSIISNSVG QTRFSGLLIV KTVLEFILQK TDSGVTLHPL VRTSKVKNEV ASFKQALSNL ARHGEYAPFA RVLNLSGINN LEHGLYPQLS AIALGVATAH GSTLAGVNVG EQYQQLREAA HDAEVKLQRR HEHQEIQAIA EDDEERKILE QFHLQKTEIT HSQTLAVLSQ KREKLARLAA EIENNIVEDQ GFKQSQNRVS QSFLNDPTPV EVTVQARPMN RVEHHHHHHH H UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (5'-R(*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3')
| Macromolecule | Name: RNA (5'-R(*AP*GP*AP*CP*AP*CP*AP*CP*AP*AP*AP*AP*AP*CP*AP*AP*GP*A)-3') type: rna / ID: 2 |
|---|---|
| Source (natural) | Organism:  Marburg virus - Musoke, Kenya, 1980 Marburg virus - Musoke, Kenya, 1980 |
| Sequence | String: AGACACACAA AAACAAGA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1.25 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 5518 / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)