+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5052 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of Abeta(1-42) amyloid fibrils | |||||||||
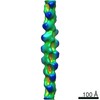
 Map data Map data | This is a cryoEM map of a segment of Abeta(1-42) amyloid fibrils. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Alzheimer's disease / amyloid fibrils / Abeta(1-42) / neurodegenerative disease / IHRSR | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Zhang R / Hu X / Khant H / Ludtke SJ / Chiu W / Schmid MF / Frieden C / Lee J-M | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: Interprotofilament interactions between Alzheimer's Abeta1-42 peptides in amyloid fibrils revealed by cryoEM. Authors: Rui Zhang / Xiaoyan Hu / Htet Khant / Steven J Ludtke / Wah Chiu / Michael F Schmid / Carl Frieden / Jin-Moo Lee /  Abstract: Alzheimer's disease is a neurodegenerative disorder characterized by the accumulation of amyloid plaques in the brain. This amyloid primarily contains amyloid-beta (Abeta), a 39- to 43-aa peptide ...Alzheimer's disease is a neurodegenerative disorder characterized by the accumulation of amyloid plaques in the brain. This amyloid primarily contains amyloid-beta (Abeta), a 39- to 43-aa peptide derived from the proteolytic cleavage of the endogenous amyloid precursor protein. The 42-residue-length Abeta peptide (Abeta(1-42)), the most abundant Abeta peptide found in plaques, has a much greater propensity to self-aggregate into fibrils than the other peptides and is believed to be more pathogenic. Synthetic human Abeta(1-42) peptides self-aggregate into stable but poorly-ordered helical filaments. We determined their structure to approximately 10-A resolution by using cryoEM and the iterative real-space reconstruction method. This structure reveals 2 protofilaments winding around a hollow core. Previous hairpin-like NMR models for Abeta(17-42) fit well in the cryoEM density map and reveal that the juxtaposed protofilaments are joined via the N terminus of the peptide from 1 protofilament connecting to the loop region of the peptide in the opposite protofilament. This model of mature Abeta(1-42) fibrils is markedly different from previous cryoEM models of Abeta(1-40) fibrils. In our model, the C terminus of Abeta forms the inside wall of the hollow core, which is supported by partial proteolysis analysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5052.map.gz emd_5052.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5052-v30.xml emd-5052-v30.xml emd-5052.xml emd-5052.xml | 11 KB 11 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5052_1.png emd_5052_1.png | 56 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5052 http://ftp.pdbj.org/pub/emdb/structures/EMD-5052 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5052 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5052 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5052.map.gz / Format: CCP4 / Size: 89 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5052.map.gz / Format: CCP4 / Size: 89 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a cryoEM map of a segment of Abeta(1-42) amyloid fibrils. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Abeta(1-42) amyloid fibril
| Entire | Name: Abeta(1-42) amyloid fibril |
|---|---|
| Components |
|
-Supramolecule #1000: Abeta(1-42) amyloid fibril
| Supramolecule | Name: Abeta(1-42) amyloid fibril / type: sample / ID: 1000 Details: The sample was prepared in 10mM HCl buffer for 1 month to form fibrils. Number unique components: 1 |
|---|
-Macromolecule #1: Abeta(1-42) amyloid fibril
| Macromolecule | Name: Abeta(1-42) amyloid fibril / type: protein_or_peptide / ID: 1 / Name.synonym: Abeta(1-42) amyloid fibril Details: The component of this amyloid fibril is a 42-residue peptide. Dry synthetic human Abeta1-42 peptides were purchased from American Peptide Company Inc. Oligomeric state: helical / Recombinant expression: No / Database: NCBI |
|---|---|
| Molecular weight | Experimental: 5 KDa / Theoretical: 5 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 2 / Details: 10mM HCl, 2% DMSO |
| Grid | Details: Quantifoil 200 mesh grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 90 K / Instrument: OTHER / Details: Vitrification instrument: vitrobot / Method: 2 blots, each 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Temperature | Min: 93 K / Max: 93 K / Average: 93 K |
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 400,000 times magnification |
| Specialist optics | Energy filter - Name: JEOL |
| Date | Dec 9, 2006 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Number real images: 208 / Average electron dose: 18 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The long straight filaments were boxed out from the raw electron micrograph using EMAN helixboxer program. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Axial symmetry: C2 (2 fold cyclic) Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 10.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IHRSR Details: We sorted heterogeneous dataset into homogeneous subgroups. And the deposited map is the reconstruction for one of the subgroups . |
| CTF correction | Details: each micrograph by binnary phase flipping |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Details | PDBEntryID_givenInChain. Protocol: manually fit. manually fit 10 abeta(1-42) monomers into one strand of cryoEM map. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















