+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
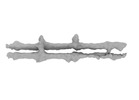
| Title | PomX filament from Myxococcus xanthus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | prokaryotes / intermediate filaments / PROTEIN FIBRIL | |||||||||
| Biological species |  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) | |||||||||
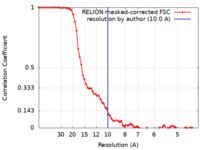
| Method | single particle reconstruction / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Xu P / Schumacher D / Liu C / Dickmanns M / Beck F / Plitzko J / Baumeister W / Sogaard-Andersen L | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: In situ architecture of a nucleoid-associated biomolecular co-condensate that regulates bacterial cell division. Authors: Peng Xu / Dominik Schumacher / Chuan Liu / Andrea Harms / Marcel Dickmanns / Florian Beck / Jürgen M Plitzko / Wolfgang Baumeister / Lotte Søgaard-Andersen /  Abstract: In most bacteria, cell division depends on the tubulin-homolog FtsZ that polymerizes in a GTP-dependent manner to form the cytokinetic Z-ring at the future division site. Subsequently, the Z-ring ...In most bacteria, cell division depends on the tubulin-homolog FtsZ that polymerizes in a GTP-dependent manner to form the cytokinetic Z-ring at the future division site. Subsequently, the Z-ring recruits, directly or indirectly, all other proteins of the divisome complex that executes cytokinesis. A critical step in this process is the precise positioning of the Z-ring at the future division site. While the divisome proteins are generally conserved, the regulatory systems that position the Z-ring are more diverse. However, these systems have in common that they modulate FtsZ polymerization. In PomX, PomY, and PomZ form precisely one MDa-sized, nonstoichiometric, nucleoid-associated assembly that spatiotemporally guides Z-ring formation. Here, using cryo-correlative light and electron microscopy together with in situ cryoelectron tomography, we determine the PomXYZ assembly's architecture at close-to-live conditions. PomX forms a porous meshwork of randomly intertwined filaments. Templated by this meshwork, the phase-separating PomY protein forms a biomolecular condensate that compacts and bends the PomX filaments, resulting in the formation of a selective PomXYZ co-condensate that is associated to the nucleoid by PomZ. These studies reveal a hitherto undescribed supramolecular structure and provide a framework for understanding how a nonstoichiometric co-condensate forms, maintains number control, and nucleates GTP-dependent FtsZ polymerization to precisely regulate cell division. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50515.map.gz emd_50515.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50515-v30.xml emd-50515-v30.xml emd-50515.xml emd-50515.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50515_fsc.xml emd_50515_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_50515.png emd_50515.png | 16.4 KB | ||
| Masks |  emd_50515_msk_1.map emd_50515_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50515.cif.gz emd-50515.cif.gz | 4.8 KB | ||
| Others |  emd_50515_half_map_1.map.gz emd_50515_half_map_1.map.gz emd_50515_half_map_2.map.gz emd_50515_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50515 http://ftp.pdbj.org/pub/emdb/structures/EMD-50515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50515 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50515 | HTTPS FTP |
-Validation report
| Summary document |  emd_50515_validation.pdf.gz emd_50515_validation.pdf.gz | 678 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50515_full_validation.pdf.gz emd_50515_full_validation.pdf.gz | 677.6 KB | Display | |
| Data in XML |  emd_50515_validation.xml.gz emd_50515_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_50515_validation.cif.gz emd_50515_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50515 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50515 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50515 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50515 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50515.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50515.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.18 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50515_msk_1.map emd_50515_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_50515_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_50515_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : filaments of PomX
| Entire | Name: filaments of PomX |
|---|---|
| Components |
|
-Supramolecule #1: filaments of PomX
| Supramolecule | Name: filaments of PomX / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 20mM Tris/HCl; 200mM NaCl; 1mM EDTA; pH 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | To purify PomX-His6 plasmid pEMR3 was propagated in E. coli(DE3) cells (NEB). Cells were grown in LB medium with kanamycin at 30 degree to an OD600 of 0.6. Protein accumulation was induced with 0.5mM IPTG for 16 hrs at 18 degree. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 17653 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)