+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
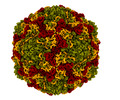
| Title | Coxsackievirus A9 bound with compound 18 (CL304) | ||||||||||||
 Map data Map data | Flipped-handedness map of Coxsackievirus A9 bound to CL304 (compound 18) resolved to 2.6 A. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Antiviral / capsid stabilizer / hydrophobic pocket / cryoEM / VIRUS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Coxsackievirus A9 / Coxsackievirus A9 /  Human coxsackievirus A9 (strain Griggs) Human coxsackievirus A9 (strain Griggs) | ||||||||||||
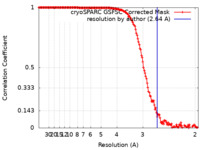
| Method | single particle reconstruction / cryo EM / Resolution: 2.64 Å | ||||||||||||
 Authors Authors | Plavec Z / Butcher SJ / Mitchell C / Buckner C | ||||||||||||
| Funding support |  Finland, 3 items Finland, 3 items
| ||||||||||||
 Citation Citation | Journal: Protein Sci / Year: 2021 Title: UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Authors: Eric F Pettersen / Thomas D Goddard / Conrad C Huang / Elaine C Meng / Gregory S Couch / Tristan I Croll / John H Morris / Thomas E Ferrin /   Abstract: UCSF ChimeraX is the next-generation interactive visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX brings (a) ...UCSF ChimeraX is the next-generation interactive visualization program from the Resource for Biocomputing, Visualization, and Informatics (RBVI), following UCSF Chimera. ChimeraX brings (a) significant performance and graphics enhancements; (b) new implementations of Chimera's most highly used tools, many with further improvements; (c) several entirely new analysis features; (d) support for new areas such as virtual reality, light-sheet microscopy, and medical imaging data; (e) major ease-of-use advances, including toolbars with icons to perform actions with a single click, basic "undo" capabilities, and more logical and consistent commands; and (f) an app store for researchers to contribute new tools. ChimeraX includes full user documentation and is free for noncommercial use, with downloads available for Windows, Linux, and macOS from https://www.rbvi.ucsf.edu/chimerax. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50414.map.gz emd_50414.map.gz | 170.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50414-v30.xml emd-50414-v30.xml emd-50414.xml emd-50414.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50414_fsc.xml emd_50414_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_50414.png emd_50414.png | 130.6 KB | ||
| Filedesc metadata |  emd-50414.cif.gz emd-50414.cif.gz | 7.7 KB | ||
| Others |  emd_50414_half_map_1.map.gz emd_50414_half_map_1.map.gz emd_50414_half_map_2.map.gz emd_50414_half_map_2.map.gz | 319.8 MB 319.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50414 http://ftp.pdbj.org/pub/emdb/structures/EMD-50414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50414 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50414 | HTTPS FTP |
-Validation report
| Summary document |  emd_50414_validation.pdf.gz emd_50414_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50414_full_validation.pdf.gz emd_50414_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_50414_validation.xml.gz emd_50414_validation.xml.gz | 24.2 KB | Display | |
| Data in CIF |  emd_50414_validation.cif.gz emd_50414_validation.cif.gz | 31.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50414 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50414 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50414 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50414 | HTTPS FTP |
-Related structure data
| Related structure data |  9fgnMC  8s7jC  9exiC  9fa9C  9fczC  9fo2C  9fo5C  9fp5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50414.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50414.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Flipped-handedness map of Coxsackievirus A9 bound to CL304 (compound 18) resolved to 2.6 A. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
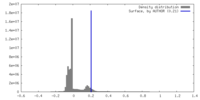
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map of Coxsackievirus A9 bound to CL304 (compound 18)
| File | emd_50414_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of Coxsackievirus A9 bound to CL304 (compound 18) | ||||||||||||
| Projections & Slices |
| ||||||||||||
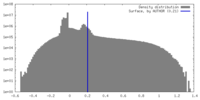
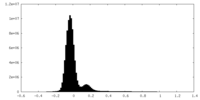
| Density Histograms |
-Half map: Half-map of Coxsackievirus A9 bound to CL304 (compound 18).
| File | emd_50414_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of Coxsackievirus A9 bound to CL304 (compound 18). | ||||||||||||
| Projections & Slices |
| ||||||||||||
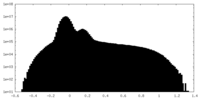
| Density Histograms |
- Sample components
Sample components
-Entire : Human coxsackievirus A9 (strain Griggs)
| Entire | Name:  Human coxsackievirus A9 (strain Griggs) Human coxsackievirus A9 (strain Griggs) |
|---|---|
| Components |
|
-Supramolecule #1: Human coxsackievirus A9 (strain Griggs)
| Supramolecule | Name: Human coxsackievirus A9 (strain Griggs) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Coxsackievirus A9 was propagated on green monkey kidney cells and purified on a sucrose gradient. NCBI-ID: 12068 / Sci species name: Human coxsackievirus A9 (strain Griggs) / Sci species strain: Griggs / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8 MDa |
| Virus shell | Shell ID: 1 / Name: icosahedral capsid / Diameter: 300.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A9 / Strain: Griggs / Tissue: kidney Coxsackievirus A9 / Strain: Griggs / Tissue: kidney |
| Molecular weight | Theoretical: 31.952896 KDa |
| Sequence | String: GDVEEAIERA VVHVADTMRS GPSNSASVPA LTAVETGHTS QVTPSDTMQT RHVKNYHSRS ESTVENFLGR SACVYMEEYK TTDNDVNKK FVAWPINTKQ MVQMRRKLEM FTYLRFDMEV TFVITSRQDP GTTLAQDMPV LTHQIMYVPP GGPIPAKVDD Y AWQTSTNP ...String: GDVEEAIERA VVHVADTMRS GPSNSASVPA LTAVETGHTS QVTPSDTMQT RHVKNYHSRS ESTVENFLGR SACVYMEEYK TTDNDVNKK FVAWPINTKQ MVQMRRKLEM FTYLRFDMEV TFVITSRQDP GTTLAQDMPV LTHQIMYVPP GGPIPAKVDD Y AWQTSTNP SIFWTEGNAP ARMSIPFISI GNAYSNFYDG WSNFDQRGSY GYNTLNNLGH IYVRHVSGSS PHPITSTIRV YF KPKHTRA WVPRPPRLCQ YKKAFSVDFT PTPITDTRKD INTVT UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A9 / Strain: Griggs / Tissue: kidney Coxsackievirus A9 / Strain: Griggs / Tissue: kidney |
| Molecular weight | Theoretical: 27.720285 KDa |
| Sequence | String: SDRVRSITLG NSTITTQECA NVVVGYGRWP TYLRDDEATA EDQPTQPDVA TCRFYTLDSI KWEKGSVGWW WKFPEALSDM GLFGQNMQY HYLGRAGYTI HVQCNASKFH QGCLLVVCVP EAEMGGAVVG QAFSATAMAN GDKAYEFTSA TQSDQTKVQT A IHNAGMGV ...String: SDRVRSITLG NSTITTQECA NVVVGYGRWP TYLRDDEATA EDQPTQPDVA TCRFYTLDSI KWEKGSVGWW WKFPEALSDM GLFGQNMQY HYLGRAGYTI HVQCNASKFH QGCLLVVCVP EAEMGGAVVG QAFSATAMAN GDKAYEFTSA TQSDQTKVQT A IHNAGMGV GVGNLTIYPH QWINLRTNNS ATIVMPYINS VPMDNMFRHY NFTLMVIPFV KLDYADTAST YVPITVTVAP MC AEYNGLR LAQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A9 / Strain: Griggs / Tissue: kidney Coxsackievirus A9 / Strain: Griggs / Tissue: kidney |
| Molecular weight | Theoretical: 26.149889 KDa |
| Sequence | String: LPTMNTPGST QFLTSDDFQS PCALPQFDVT PSMNIPGEVK NLMEIAEVDS VVPVNNVQDT TDQMEMFRIP VTINAPLQQQ VFGLRLQPG LDSVFKHTLL GEILNYYAHW SGSMKLTFVF CGSAMATGKF LIAYSPPGAN PPKTRKDAML GTHIIWDIGL Q SSCVLCVP ...String: LPTMNTPGST QFLTSDDFQS PCALPQFDVT PSMNIPGEVK NLMEIAEVDS VVPVNNVQDT TDQMEMFRIP VTINAPLQQQ VFGLRLQPG LDSVFKHTLL GEILNYYAHW SGSMKLTFVF CGSAMATGKF LIAYSPPGAN PPKTRKDAML GTHIIWDIGL Q SSCVLCVP WISQTHYRLV QQDEYTSAGY VTCWYQTGMI VPPGTPNSSS IMCFASACND FSVRMLRDTP FISQDNKL UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus A9 / Strain: Griggs / Tissue: kidney Coxsackievirus A9 / Strain: Griggs / Tissue: kidney |
| Molecular weight | Theoretical: 7.237936 KDa |
| Sequence | String: GAQVSTQKTG AHETSLSAAG NSIIHYTNIN YYKDAASNSA NRQDFTQDPS KFTEPVKDVM IKSLPAL UniProtKB: Genome polyprotein |
-Macromolecule #5: ~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-4-[(4-methylpiperazin-1-y...
| Macromolecule | Name: ~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-4-[(4-methylpiperazin-1-yl)methyl]aniline type: ligand / ID: 5 / Number of copies: 1 / Formula: A1ICH |
|---|---|
| Molecular weight | Theoretical: 331.403 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Details: PBS containing 2 mM MgCl2 and 5% DMSO. |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 295 K / Instrument: LEICA EM GP Details: Sample was incubated for 15 s on the grid before blotted from the front for 1.5 s.. |
| Details | Compound 18 (CL304) was solubilized in DMSO and diluted to the final concentration in PBS + 2 mM MgCl2. Purified Coxsackievirus A9 was incubated with compound 18 (CL304) for 1h at 37C after which the sample was vitrified on a semi-automatic plunger Leica EM GP on copper Quantifoil R1.2/1.3 grids with 2 nm thin carbon film and 300 mesh. This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 93.15 K / Max: 103.15 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 1496 pixel / Digitization - Dimensions - Height: 1496 pixel / Number real images: 625 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 150000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)