+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
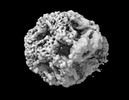
| Title | mjHSP16.5 26mer (+lysozyme, 75C) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | sHSP / thermophile / holdase / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein complex oligomerization / response to salt stress / protein folding chaperone / response to hydrogen peroxide / unfolded protein binding / protein folding / response to heat / protein stabilization / protein-containing complex / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Methanocaldococcus jannaschii (archaea) Methanocaldococcus jannaschii (archaea) | |||||||||
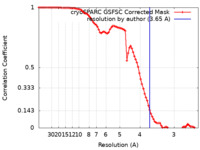
| Method | single particle reconstruction / cryo EM / Resolution: 3.65 Å | |||||||||
 Authors Authors | Miller AP / Reichow SL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Mechanism of small heat shock protein client sequestration and induced polydispersity. Authors: Adam P Miller / Steve L Reichow /  Abstract: Small heat shock proteins (sHSPs) act as first responders during cellular stress, sequestering destabilized proteins (clients) to prevent aggregation and facilitate refolding or degradation. This ...Small heat shock proteins (sHSPs) act as first responders during cellular stress, sequestering destabilized proteins (clients) to prevent aggregation and facilitate refolding or degradation. This critical function, conserved across all life, is linked to proteostasis and protein misfolding diseases. However, the extreme molecular plasticity of sHSP/client complexes has limited mechanistic understanding. Here, we present high-resolution cryo-EM structures of Methanocaldococcus jannaschii sHSP (mjHSP16.5) in apo and multiple client-bound states. The ensemble reveals molecular mechanisms of client sequestration, highlighting cooperative chaperone-client interactions. Client engagement polarizes scaffold stability, promoting higher-order assembly and enhanced sequestration. Higher-order states suggest multiple sHSP/client assembly pathways, including subunit insertion at destabilized geometrical features. These findings provide critical insights into sHSP chaperone function and the interplay between polydispersity and client handling under stress. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_49834.map.gz emd_49834.map.gz | 31.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-49834-v30.xml emd-49834-v30.xml emd-49834.xml emd-49834.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_49834_fsc.xml emd_49834_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_49834.png emd_49834.png | 63 KB | ||
| Filedesc metadata |  emd-49834.cif.gz emd-49834.cif.gz | 5.5 KB | ||
| Others |  emd_49834_half_map_1.map.gz emd_49834_half_map_1.map.gz emd_49834_half_map_2.map.gz emd_49834_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-49834 http://ftp.pdbj.org/pub/emdb/structures/EMD-49834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49834 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-49834 | HTTPS FTP |
-Related structure data
| Related structure data |  9nvfMC  9nv4C  9nv7C  9nv8C  9nvcC  9nviC  9nvjC  9nvkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_49834.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_49834.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_49834_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_49834_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mjHSP16.5 26mer
| Entire | Name: mjHSP16.5 26mer |
|---|---|
| Components |
|
-Supramolecule #1: mjHSP16.5 26mer
| Supramolecule | Name: mjHSP16.5 26mer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Methanocaldococcus jannaschii (archaea) Methanocaldococcus jannaschii (archaea) |
-Macromolecule #1: Small heat shock protein HSP16.5
| Macromolecule | Name: Small heat shock protein HSP16.5 / type: protein_or_peptide / ID: 1 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Methanocaldococcus jannaschii (archaea) Methanocaldococcus jannaschii (archaea) |
| Molecular weight | Theoretical: 12.629581 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIQISGKGFM PISIIEGDQH IKVIAWLPGV NKEDIILNAV GDTLEIRAKR SPLMITESER IIYSEIPEEE EIYRTIKLPA TVKEENASA KFENGVLSVI LPKAESSIKK GINIE UniProtKB: Small heat shock protein HSP16.5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)