+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4980 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of an MCM loading intermediate | |||||||||
 マップデータ マップデータ | Full map - complete MCM-ORC (MO) origin licensing intermediate | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | DNA Replication / Origin licensing / MCM2-7 helicase / Origin Recognition Complex / REPLICATION | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / Assembly of the ORC complex at the origin of replication / premeiotic DNA replication ...CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / Assembly of the ORC complex at the origin of replication / premeiotic DNA replication / nuclear origin of replication recognition complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / mitotic DNA replication / Activation of the pre-replicative complex / CMG complex / nuclear pre-replicative complex / nucleosome organization / Activation of ATR in response to replication stress / DNA replication preinitiation complex / MCM complex / replication fork protection complex / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / single-stranded DNA helicase activity / mitotic DNA replication initiation / regulation of DNA-templated DNA replication initiation / silent mating-type cassette heterochromatin formation / DNA strand elongation involved in DNA replication / Orc1 removal from chromatin / nuclear replication fork / regulation of DNA replication / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / nucleosome binding / helicase activity / transcription elongation by RNA polymerase II / heterochromatin formation / single-stranded DNA binding / DNA helicase / forked DNA-dependent helicase activity / single-stranded 3'-5' DNA helicase activity / four-way junction helicase activity / double-stranded DNA helicase activity / chromosome, telomeric region / DNA replication / DNA damage response / chromatin binding / ATP hydrolysis activity / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |   | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 4.4 Å | |||||||||
 データ登録者 データ登録者 | Miller TCR / Locke J | |||||||||
| 資金援助 |  英国, 2件 英国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2019 ジャーナル: Nature / 年: 2019タイトル: Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. 著者: Thomas C R Miller / Julia Locke / Julia F Greiwe / John F X Diffley / Alessandro Costa /  要旨: In preparation for bidirectional DNA replication, the origin recognition complex (ORC) loads two hexameric MCM helicases to form a head-to-head double hexamer around DNA. The mechanism of MCM double- ...In preparation for bidirectional DNA replication, the origin recognition complex (ORC) loads two hexameric MCM helicases to form a head-to-head double hexamer around DNA. The mechanism of MCM double-hexamer formation is debated. Single-molecule experiments have suggested a sequential mechanism, in which the ORC-dependent loading of the first hexamer drives the recruitment of the second hexamer. By contrast, biochemical data have shown that two rings are loaded independently via the same ORC-mediated mechanism, at two inverted DNA sites. Here we visualize MCM loading using time-resolved electron microscopy, and identify intermediates in the formation of the double hexamer. We confirm that both hexamers are recruited via the same interaction that occurs between ORC and the C-terminal domains of the MCM helicases. Moreover, we identify the mechanism of coupled MCM loading. The loading of the first MCM hexamer around DNA creates a distinct interaction site, which promotes the engagement of ORC at the N-terminal homodimerization interface of MCM. In this configuration, ORC is poised to direct the recruitment of the second hexamer in an inverted orientation, which is suitable for the formation of the double hexamer. Our results therefore reconcile the two apparently contrasting models derived from single-molecule experiments and biochemical data. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4980.map.gz emd_4980.map.gz | 9.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4980-v30.xml emd-4980-v30.xml emd-4980.xml emd-4980.xml | 62.1 KB 62.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_4980_fsc_1.xml emd_4980_fsc_1.xml emd_4980_fsc_2.xml emd_4980_fsc_2.xml emd_4980_fsc_3.xml emd_4980_fsc_3.xml | 12.1 KB 12.1 KB 12.1 KB | 表示 表示 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_4980.png emd_4980.png | 131.5 KB | ||
| マスクデータ |  emd_4980_msk_1.map emd_4980_msk_1.map emd_4980_msk_2.map emd_4980_msk_2.map emd_4980_msk_3.map emd_4980_msk_3.map | 149.9 MB 149.9 MB 149.9 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-4980.cif.gz emd-4980.cif.gz | 13.7 KB | ||
| その他 |  emd_4980_additional_1.map.gz emd_4980_additional_1.map.gz emd_4980_additional_2.map.gz emd_4980_additional_2.map.gz emd_4980_additional_3.map.gz emd_4980_additional_3.map.gz emd_4980_additional_4.map.gz emd_4980_additional_4.map.gz emd_4980_additional_5.map.gz emd_4980_additional_5.map.gz emd_4980_additional_6.map.gz emd_4980_additional_6.map.gz emd_4980_half_map_1.map.gz emd_4980_half_map_1.map.gz emd_4980_half_map_2.map.gz emd_4980_half_map_2.map.gz | 96.8 MB 96.8 MB 8.4 MB 108.5 MB 6.5 MB 108.5 MB 118.1 MB 118.2 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4980 http://ftp.pdbj.org/pub/emdb/structures/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4980 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_4980_validation.pdf.gz emd_4980_validation.pdf.gz | 414.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_4980_full_validation.pdf.gz emd_4980_full_validation.pdf.gz | 413.5 KB | 表示 | |
| XML形式データ |  emd_4980_validation.xml.gz emd_4980_validation.xml.gz | 12.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4980 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4980 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4980.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4980.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Full map - complete MCM-ORC (MO) origin licensing intermediate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
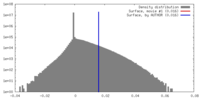
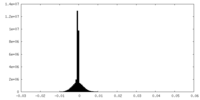
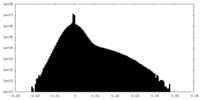
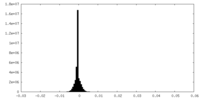
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
+マスク #1
+マスク #2
+マスク #3
+追加マップ: Half map - MCM-Orc6N lobe of MCM-ORC (MO)...
+追加マップ: Half map - MCM-Orc6N lobe of MCM-ORC (MO)...
+追加マップ: Full map - MCM-Orc6N lobe of MCM-ORC (MO)...
+追加マップ: Half map - Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+追加マップ: Full map - Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+追加マップ: Half map – Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+ハーフマップ: Half map - complete MCM-ORC (MO) origin licensing intermediate
+ハーフマップ: Half map - complete MCM-ORC (MO) origin licensing intermediate
- 試料の構成要素
試料の構成要素
+全体 : The MCM-ORC (MO) loading intermediate
+超分子 #1: The MCM-ORC (MO) loading intermediate
+超分子 #2: MCM-Orc6N lobe of the MCM-ORC (MO) origin licensing intermediate.
+超分子 #3: Orc1-5-Orc6C lobe of the MCM-ORC (MO) origin licensing intermediate
+超分子 #4: The MCM-ORC (MO) loading intermediate protein complex
+超分子 #5: DNA
+分子 #1: Origin recognition complex subunit 1
+分子 #2: Origin recognition complex subunit 2
+分子 #3: Origin recognition complex subunit 3
+分子 #4: Origin recognition complex subunit 4
+分子 #5: Origin recognition complex subunit 5
+分子 #6: Origin recognition complex subunit 6
+分子 #7: DNA replication licensing factor MCM2
+分子 #8: DNA replication licensing factor MCM3
+分子 #9: DNA replication licensing factor MCM4
+分子 #10: Minichromosome maintenance protein 5
+分子 #11: DNA replication licensing factor MCM6
+分子 #12: DNA replication licensing factor MCM7
+分子 #13: DNA (88-MER)
+分子 #14: DNA (88-MER)
+分子 #15: ADENOSINE-5'-TRIPHOSPHATE
+分子 #16: MAGNESIUM ION
+分子 #17: ADENOSINE-5'-DIPHOSPHATE
+分子 #18: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 構成要素:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| グリッド | 材質: COPPER / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: LACEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 雰囲気: AIR | ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / チャンバー内温度: 288 K / 装置: LEICA EM GP 詳細: 10 second incubation, 3.5 seconds single side blotting.. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - 画像ごとのフレーム数: 1-30 / 撮影したグリッド数: 1 / 平均電子線量: 1.68 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT | ||||||||||
| 得られたモデル |  PDB-6rqc:  PDB-9i3i: |
 ムービー
ムービー コントローラー
コントローラー

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)