+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4980 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an MCM loading intermediate | |||||||||
 Map data Map data | Full map - complete MCM-ORC (MO) origin licensing intermediate | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA Replication / Origin licensing / MCM2-7 helicase / Origin Recognition Complex / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationCDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / premeiotic DNA replication / Assembly of the ORC complex at the origin of replication ...CDC6 association with the ORC:origin complex / Cul8-RING ubiquitin ligase complex / maintenance of rDNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / MCM complex binding / nuclear DNA replication / premeiotic DNA replication / Assembly of the ORC complex at the origin of replication / replication fork protection complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / nuclear origin of replication recognition complex / Activation of the pre-replicative complex / mitotic DNA replication / CMG complex / nuclear pre-replicative complex / DNA replication preinitiation complex / Activation of ATR in response to replication stress / MCM complex / nucleosome organization / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / mitotic DNA replication initiation / single-stranded DNA helicase activity / silent mating-type cassette heterochromatin formation / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / Orc1 removal from chromatin / nuclear replication fork / regulation of DNA replication / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / nucleosome binding / DNA helicase activity / transcription elongation by RNA polymerase II / helicase activity / heterochromatin formation / single-stranded DNA binding / DNA helicase / DNA replication / chromosome, telomeric region / DNA damage response / chromatin binding / ATP hydrolysis activity / zinc ion binding / nucleoplasm / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Miller TCR / Locke J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Authors: Thomas C R Miller / Julia Locke / Julia F Greiwe / John F X Diffley / Alessandro Costa /  Abstract: In preparation for bidirectional DNA replication, the origin recognition complex (ORC) loads two hexameric MCM helicases to form a head-to-head double hexamer around DNA. The mechanism of MCM double- ...In preparation for bidirectional DNA replication, the origin recognition complex (ORC) loads two hexameric MCM helicases to form a head-to-head double hexamer around DNA. The mechanism of MCM double-hexamer formation is debated. Single-molecule experiments have suggested a sequential mechanism, in which the ORC-dependent loading of the first hexamer drives the recruitment of the second hexamer. By contrast, biochemical data have shown that two rings are loaded independently via the same ORC-mediated mechanism, at two inverted DNA sites. Here we visualize MCM loading using time-resolved electron microscopy, and identify intermediates in the formation of the double hexamer. We confirm that both hexamers are recruited via the same interaction that occurs between ORC and the C-terminal domains of the MCM helicases. Moreover, we identify the mechanism of coupled MCM loading. The loading of the first MCM hexamer around DNA creates a distinct interaction site, which promotes the engagement of ORC at the N-terminal homodimerization interface of MCM. In this configuration, ORC is poised to direct the recruitment of the second hexamer in an inverted orientation, which is suitable for the formation of the double hexamer. Our results therefore reconcile the two apparently contrasting models derived from single-molecule experiments and biochemical data. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4980.map.gz emd_4980.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4980-v30.xml emd-4980-v30.xml emd-4980.xml emd-4980.xml | 62.1 KB 62.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4980_fsc_1.xml emd_4980_fsc_1.xml emd_4980_fsc_2.xml emd_4980_fsc_2.xml emd_4980_fsc_3.xml emd_4980_fsc_3.xml | 12.1 KB 12.1 KB 12.1 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_4980.png emd_4980.png | 131.5 KB | ||
| Masks |  emd_4980_msk_1.map emd_4980_msk_1.map emd_4980_msk_2.map emd_4980_msk_2.map emd_4980_msk_3.map emd_4980_msk_3.map | 149.9 MB 149.9 MB 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4980.cif.gz emd-4980.cif.gz | 13.7 KB | ||
| Others |  emd_4980_additional_1.map.gz emd_4980_additional_1.map.gz emd_4980_additional_2.map.gz emd_4980_additional_2.map.gz emd_4980_additional_3.map.gz emd_4980_additional_3.map.gz emd_4980_additional_4.map.gz emd_4980_additional_4.map.gz emd_4980_additional_5.map.gz emd_4980_additional_5.map.gz emd_4980_additional_6.map.gz emd_4980_additional_6.map.gz emd_4980_half_map_1.map.gz emd_4980_half_map_1.map.gz emd_4980_half_map_2.map.gz emd_4980_half_map_2.map.gz | 96.8 MB 96.8 MB 8.4 MB 108.5 MB 6.5 MB 108.5 MB 118.1 MB 118.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4980 http://ftp.pdbj.org/pub/emdb/structures/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4980 | HTTPS FTP |
-Related structure data
| Related structure data |  6rqcMC  9i3iM M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4980.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4980.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map - complete MCM-ORC (MO) origin licensing intermediate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
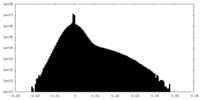
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
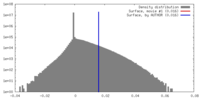
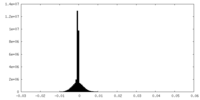
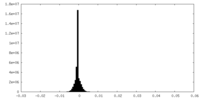
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Mask #3
+Additional map: Half map - MCM-Orc6N lobe of MCM-ORC (MO)...
+Additional map: Half map - MCM-Orc6N lobe of MCM-ORC (MO)...
+Additional map: Full map - MCM-Orc6N lobe of MCM-ORC (MO)...
+Additional map: Half map - Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+Additional map: Full map - Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+Additional map: Half map – Orc1-5-Orc6C lobe of MCM-ORC (MO)...
+Half map: Half map - complete MCM-ORC (MO) origin licensing intermediate
+Half map: Half map - complete MCM-ORC (MO) origin licensing intermediate
- Sample components
Sample components
+Entire : The MCM-ORC (MO) loading intermediate
+Supramolecule #1: The MCM-ORC (MO) loading intermediate
+Supramolecule #2: MCM-Orc6N lobe of the MCM-ORC (MO) origin licensing intermediate.
+Supramolecule #3: Orc1-5-Orc6C lobe of the MCM-ORC (MO) origin licensing intermediate
+Supramolecule #4: The MCM-ORC (MO) loading intermediate protein complex
+Supramolecule #5: DNA
+Macromolecule #1: Origin recognition complex subunit 1
+Macromolecule #2: Origin recognition complex subunit 2
+Macromolecule #3: Origin recognition complex subunit 3
+Macromolecule #4: Origin recognition complex subunit 4
+Macromolecule #5: Origin recognition complex subunit 5
+Macromolecule #6: Origin recognition complex subunit 6
+Macromolecule #7: DNA replication licensing factor MCM2
+Macromolecule #8: DNA replication licensing factor MCM3
+Macromolecule #9: DNA replication licensing factor MCM4
+Macromolecule #10: Minichromosome maintenance protein 5
+Macromolecule #11: DNA replication licensing factor MCM6
+Macromolecule #12: DNA replication licensing factor MCM7
+Macromolecule #13: DNA (88-MER)
+Macromolecule #14: DNA (88-MER)
+Macromolecule #15: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #16: MAGNESIUM ION
+Macromolecule #17: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #18: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 288 K / Instrument: LEICA EM GP Details: 10 second incubation, 3.5 seconds single side blotting.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-30 / Number grids imaged: 1 / Average electron dose: 1.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||||||
| Output model |  PDB-6rqc:  PDB-9i3i: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)