+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4917 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the dynein-2 complex; motor domains | |||||||||||||||||||||||||||
 Map data Map data | Motor domain map, sharpened and masked | |||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | dynein / cilia / intraflagellar transport / complex / motor protein | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmethylated-DNA-[protein]-cysteine S-methyltransferase / methylated-DNA-[protein]-cysteine S-methyltransferase activity / cell projection organization / minus-end-directed microtubule motor activity / dynein light intermediate chain binding / cytoplasmic dynein complex / microtubule-based movement / dynein intermediate chain binding / axoneme / methylation ...methylated-DNA-[protein]-cysteine S-methyltransferase / methylated-DNA-[protein]-cysteine S-methyltransferase activity / cell projection organization / minus-end-directed microtubule motor activity / dynein light intermediate chain binding / cytoplasmic dynein complex / microtubule-based movement / dynein intermediate chain binding / axoneme / methylation / microtubule / DNA repair / DNA binding / ATP binding / metal ion binding / nucleus / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
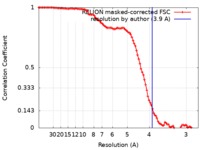
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||||||||||||||||||||
 Authors Authors | Toropova K / Zalyte R | |||||||||||||||||||||||||||
| Funding support |  United Kingdom, 8 items United Kingdom, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Authors: Katerina Toropova / Ruta Zalyte / Aakash G Mukhopadhyay / Miroslav Mladenov / Andrew P Carter / Anthony J Roberts /  Abstract: Dynein-2 assembles with polymeric intraflagellar transport (IFT) trains to form a transport machinery that is crucial for cilia biogenesis and signaling. Here we recombinantly expressed the ~1.4-MDa ...Dynein-2 assembles with polymeric intraflagellar transport (IFT) trains to form a transport machinery that is crucial for cilia biogenesis and signaling. Here we recombinantly expressed the ~1.4-MDa human dynein-2 complex and solved its cryo-EM structure to near-atomic resolution. The two identical copies of the dynein-2 heavy chain are contorted into different conformations by a WDR60-WDR34 heterodimer and a block of two RB and six LC8 light chains. One heavy chain is steered into a zig-zag conformation, which matches the periodicity of the anterograde IFT-B train. Contacts between adjacent dyneins along the train indicate a cooperative mode of assembly. Removal of the WDR60-WDR34-light chain subcomplex renders dynein-2 monomeric and relieves autoinhibition of its motility. Our results converge on a model in which an unusual stoichiometry of non-motor subunits controls dynein-2 assembly, asymmetry, and activity, giving mechanistic insight into the interaction of dynein-2 with IFT trains and the origin of diverse functions in the dynein family. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4917.map.gz emd_4917.map.gz | 6.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4917-v30.xml emd-4917-v30.xml emd-4917.xml emd-4917.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4917_fsc.xml emd_4917_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_4917.png emd_4917.png | 137.6 KB | ||
| Masks |  emd_4917_msk_1.map emd_4917_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4917.cif.gz emd-4917.cif.gz | 9 KB | ||
| Others |  emd_4917_additional.map.gz emd_4917_additional.map.gz emd_4917_half_map_1.map.gz emd_4917_half_map_1.map.gz emd_4917_half_map_2.map.gz emd_4917_half_map_2.map.gz | 64 MB 64.2 MB 64.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4917 http://ftp.pdbj.org/pub/emdb/structures/EMD-4917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4917 | HTTPS FTP |
-Related structure data
| Related structure data |  6rlaMC  4918C  6rlbC  6sc2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4917.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4917.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Motor domain map, sharpened and masked | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4917_msk_1.map emd_4917_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Motor domain map, unsharpened and unmasked
| File | emd_4917_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Motor domain map, unsharpened and unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Motor domain half map 2, unmasked
| File | emd_4917_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Motor domain half map 2, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Motor domain half map 1, unmasked
| File | emd_4917_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Motor domain half map 1, unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dynein-2 complex; motor domains
| Entire | Name: Dynein-2 complex; motor domains |
|---|---|
| Components |
|
-Supramolecule #1: Dynein-2 complex; motor domains
| Supramolecule | Name: Dynein-2 complex; motor domains / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: O6-alkylguanine-DNA alkyltransferase mutant,DYNC2H1 variant protein
| Macromolecule | Name: O6-alkylguanine-DNA alkyltransferase mutant,DYNC2H1 variant protein type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 515.223031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GDKDCEMKRT TLDSPLGKLE LSGCEQGLHR IIFLGKGTSA ADAVEVPAPA AVLGGPEPLM QATAWLNAYF HQPEAIEEFP VPALHHPVF QQESFTRQVL WKLLKVVKFG EVISYSHLAA LAGNPAATAA VKTALSGNPV PILIPCHRVV QGDLDVGGYE G GLAVKEWL ...String: GDKDCEMKRT TLDSPLGKLE LSGCEQGLHR IIFLGKGTSA ADAVEVPAPA AVLGGPEPLM QATAWLNAYF HQPEAIEEFP VPALHHPVF QQESFTRQVL WKLLKVVKFG EVISYSHLAA LAGNPAATAA VKTALSGNPV PILIPCHRVV QGDLDVGGYE G GLAVKEWL LAHEGHRLGK PGLGGSLEVL FQGPDYDIPT TLEVLFQGPA NGTADVRKLF IFTTTQNYFG LMSELWDQPL LC NCLEINN FLDDGNQMLL RVQRSDAGIS FSNTIEFGDT KDKVLVFFKL RPEVITDENL HDNILVSSML ESPISSLYQA VRQ VFAPML LKDQEWSRNF DPKLQNLLSE LEAGLGIVLR RSDTNLTKLK FKEDDTRGIL TPSDEFQFWI EQAHRGNKQI SKER ANYFK ELFETIAREF YNLDSLSLLE VVDLVETTQD VVDDVWRQTE HDHYPESRML HLLDIIGGSF GRFVQKKLGT LNLWE DPYY LVKESLKAGI SICEQWVIVC NHLTGQVWQR YVPHPWKNEK YFPETLDKLG KRLEEVLAIR TIHEKFLYFL PASEEK IIC LTRVFEPFTG LNPVQYNPYT EPLWKAAVSQ YEKIIAPAEQ KIAGKLKNYI SEIQDSPQQL LQAFLKYKEL VKRPTIS KE LMLERETLLA RLVDSIKDFR LDFENRCRGI PGDASGPLSG KNLSEVVNSI VWVRQLELKV DDTIKIAEAL LSDLPGFR C FHQSAKDLLD QLKLYEQEQF DDWSRDIQSG LSDSRSGLCI EASSRIMELD SNDGLLKVHY SDRLVILLRE VRQLSALGF VIPAKIQQVA NIAQKFCKQA IILKQVAHFY NSIDQQMIQS QRPMMLQSAL AFEQIIKNSK AGSGGKSQIT WDNPKELEGY IQKLQNAAE RLATENRKLR KWHTTFCEKV VVLMNIDLLR QQQRWKDGLQ ELRTGLATVE AQGFQASDMH AWKQHWNHQL Y KALEHQYQ MGLEALNENL PEINIDLTYK QGRLQFRPPF EEIRAKYYRE MKRFIGIPNQ FKGVGEAGDE SIFSIMIDRN AS GFLTIFS KAEDLFRRLS AVLHQHKEWI VIGQVDMEAL VEKHLFTVHD WEKNFKALKI KGKEVERLPS AVKVDCLNIN CNP VKTVID DLIQKLFDLL VLSLKKSIQA HLHEIDTFVT EAMEVLTIMP QSVEEIGDAN LQYSKLQERK PEILPLFQEA EDKN RLLRT VAGGGLETIS NLKAKWDKFE LMMESHQLMI KDQIEVMKGN VKSRLQIYYQ ELEKFKARWD QLKPGDDVIE TGQHN TLDK SAKLIKEKKI EFDDLEVTRK KLVDDCHHFR LEEPNFSLAS SISKDIESCA QIWAFYEEFQ QGFQEMANED WITFRT KTY LFEEFLMNWH DRLRKVEEHS VMTVKLQSEV DKYKIVIPIL KYVRGEHLSP DHWLDLFRLL GLPRGTSLEK LLFGDLL RV ADTIVAKAAD LKDLNSRAQG EVTIREALRE LDLWGVGAVF TLIDYEDSQS RTMKLIKDWK DIVNQVGDNR CLLQSLKD S PYYKGFEDKV SIWERKLAEL DEYLQNLNHI QRKWVYLEPI FGRGALPKEQ TRFNRVDEDF RSIMTDIKKD NRVTTLTTH AGIRNSLLTI LDQLQRCQRS LNEFLEEKRS AFPRFYFIGD DDLLEILGQS TNPSVIQSHL KKLFAGINSV CFDEKSKHIT AMKSLEGEV VPFKNKVPLS NNVETWLNDL ALEMKKTLEQ LLKECVTTGR SSQGAVDPSL FPSQILCLAE QIKFTEDVEN A IKDHSLHQ IETQLVNKLE QYTNIDTSSE DPGNTESGIL ELKLKALILD IIHNIDVVKQ LNQIQVHTTE DWAWKKQLRF YM KSDHTCC VQMVDSEFQY TYEYQGNASK LVYTPLTDKC YLTLTQAMKM GLGGNPYGPA GTGKTESVKA LGGLLGRQVL VFN CDEGID VKSMGRIFVG LVKCGAWGCF DEFNRLEESV LSAVSMQIQT IQDALKNHRT VCELLGKEVE VNSNSGIFIT MNPA GKGYG GRQKLPDNLK QLFRPVAMSH PDNELIAEVI LYSEGFKDAK VLSRKLVAIF NLSRELLTPQ QHYDWGLRAL KTVLR GSGN LLRQLNKSGT TQNANESHIV VQALRLNTMS KFTFTDCTRF DALIKDVFPG IELKEVEYDE LSAALKQVFE EANYEI IPN QIKKALELYE QLCQRMGVVI VGPSGAGKST LWRMLRAALC KTGKVVKQYT MNPKAMPRYQ LLGHIDMDTR EWSDGVL TN SARQVVREPQ DVSSWIICDG DIDPEWIESL NSVLDDNRLL TMPSGERIQF GPNVNFVFET HDLSCASPAT ISRMGMIF L SDEETDLNSL IKSWLRNQPA EYRNNLENWI GDYFEKALQW VLKQNDYVVE TSLVGTVMNG LSHLHGCRDH DEFIINLIR GLGGNLNMKS RLEFTKEVFH WARESPPDFH KPMDTYYDST RGRLATYVLK KPEDLTADDF SNGLTLPVIQ TPDMQRGLDY FKPWLSSDT KQPFILVGPE GCGKGMLLRY AFSQLRSTQI ATVHCSAQTT SRHLLQKLSQ TCMVISTNTG RVYRPKDCER L VLYLKDIN LPKLDKWGTS TLVAFLQQVL TYQGFYDENL EWVGLENIQI VASMSAGGRL GRHKLTTRFT SIVRLCSIDY PE REQLQTI YGAYLEPVLH KNLKNHSIWG SSSKIYLLAG SMVQVYEQVR AKFTVDDYSH YFFTPCILTQ WVLGLFRYDL EGG SSNHPL DYVLEIVAYE ARRLFRDKIV GAKELHLFDI ILTSVFQGDW GSDILDNMSD SFYVTWGARH NSGARAAPGQ PLPP HGKPL GKLNSTDLKD VIKKGLIHYG RDNQNLDILL FHEVLEYMSR IDRVLSFPGG SLLLAGRSGV GRRTITSLVS HMHGA VLFS PKISRGYELK QFKNDLKHVL QLAGIEAQQV VLLLEDYQFV HPTFLEMINS LLSSGEVPGL YTLEELEPLL LPLKDQ ASQ DGFFGPVFNY FTYRIQQNLH IVLIMDSANS NFMINCESNP ALHKKCQVLW MEGWSNSSMK KIPEMLFSET GGGEKYN DK KRKEEKKKNS VDPDFLKSFL LIHESCKAYG ATPSQYMTFL HVYSAISSSK KKELLKRQSH LQAGVSKLNE AKALVDEL N RKAGEQSVLL KTKQDEADAA LQMITVSMQD ASEQKTELER LKHRIAEEVV KIEERKNKID DELKEVQPLV NEAKLAVGN IKPESLSEIR SLRMPPDVIR DILEGVLRLM GIFDTSWVSM KSFLAKRGVR EDIATFDARN ISKEIRESVE ELLFKNKGSF DPKNAKRAS TAAAPLAAWV KANIQYSHVL ERIHPLETEQ AGLESNLKKT EDRKRKLEEL LNSVGQKVSE LKEKFQSRTS E AAKLEAEV SKAQETIKAA EVLINQLDRE HKRWNAQVVE ITEELATLPK RAQLAAAFIT YLSAAPESLR KTCLEEWTKS AG LEKFDLR RFLCTESEQL IWKSEGLPSD DLSIENALVI LQSRVCPFLI DPSSQATEWL KTHLKDSRLE VINQQDSNFI TAL ELAVRF GKTLIIQEMD GVEPVLYPLL RRDLVAQGPR YVVQIGDKII DYNEEFRLFL STRNPNPFIP PDAASIVTEV NFTT TRSGL RGQLLALTIQ HEKPDLEEQK TKLLQQEEDK KIQLAKLEES LLETLATSQG NILENKDLIE SLNQTKASSA LIQES LKES YKLQISLDQE RDAYLPLAES ASKMYFIISD LSKINNMYRF SLAAFLRLFQ RALQNKQDSE NTEQRIQSLI SSLQHM VYE YICRCLFKAD QLMFALHFVR GMHPELFQEN EWDTFTGVVV GDMLRKADSQ QKIRDQLPSW IDQERSWAVA TLKIALP SL YQTLCFEDAA LWRTYYNNSM CEQEFPSILA KKVSLFQQIL VVQVLRPDRL QSAMALFACK TLGLKEVSPL PLNLKRLY K ETLEIEPILI IISPGADPSQ ELQELANAER SGECYHQVAM GQGQADLAIQ MLKECARNGD WLCLKNLHLV VSWLPVLEK ELNTLQPKDT FRLWLTAEVH PNFTPILLQS SLKITYESPP GLKKNLMRTY ESWTPEQISK KDNTHRAHAL FSLAWFHAAC QERRNYIPQ GWTKFYEFSL SDLRAGYNII DRLFDGAKDV QWEFVHGLLE NAIYGGRIDN YFDLRVLQSY LKQFFNSSVI D VFNQRNKK SIFPYSVSLP QSCSILDYRA VIEKIPEDDK PSFFGLPANI ARSSQRMISS QVISQLRILG RSITAGSKFD RE IWSNELS PVLNLWKKLN QNSNLIHQKV PPPNDRQGSP ILSFIILEQF NAIRLVQSVH QSLAALSKVI RGTTLLSSEV QKL ASALLN QKCPLAWQSK WEGPEDPLQY LRGLVARALA IQNWVDKAEK QALLSETLDL SELFHPDTFL NALRQETARA VGRS VDSLK FVASWKGRLQ EAKLQIKISG LLLEGCSFDG NQLSENQLDS PSVSSVLPCF MGWIPQDACG PYSPDECISL PVYTS AERD RVVTNIDVPC GGNQDQWIQC GAALFLKNQ UniProtKB: Methylated-DNA--protein-cysteine methyltransferase, Cytoplasmic dynein 2 heavy chain 1 |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 49.6 e/Å2 Details: Average electron dose per image (e-/A2) for additional datasets was 46.8 and 45.4 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)