[English] 日本語
 Yorodumi
Yorodumi- EMDB-4909: E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4909 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | ||||||||||||
 Map data Map data | E. coli DNA Gyrase DNA binding and cleavage domain. The TOPRIM insertion was masked away during refinement. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Isomerase / Complex / DNA Gyrase / Inhibitor / DNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of DNA-templated DNA replication / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex / DNA negative supercoiling activity / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / ATP-dependent activity, acting on DNA / DNA-templated DNA replication / chromosome / response to xenobiotic stimulus ...negative regulation of DNA-templated DNA replication / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex / DNA negative supercoiling activity / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / ATP-dependent activity, acting on DNA / DNA-templated DNA replication / chromosome / response to xenobiotic stimulus / response to antibiotic / DNA-templated transcription / DNA binding / ATP binding / metal ion binding / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |    | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | ||||||||||||
 Authors Authors | Vanden Broeck A / Lamour V | ||||||||||||
| Funding support |  France, 3 items France, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex. Authors: Arnaud Vanden Broeck / Christophe Lotz / Julio Ortiz / Valérie Lamour /  Abstract: DNA gyrase is an essential enzyme involved in the homeostatic control of DNA supercoiling and the target of successful antibacterial compounds. Despite extensive studies, a detailed architecture of ...DNA gyrase is an essential enzyme involved in the homeostatic control of DNA supercoiling and the target of successful antibacterial compounds. Despite extensive studies, a detailed architecture of the full-length DNA gyrase from the model organism E. coli is still missing. Herein, we report the complete structure of the E. coli DNA gyrase nucleoprotein complex trapped by the antibiotic gepotidacin, using phase-plate single-particle cryo-electron microscopy. Our data unveil the structural and spatial organization of the functional domains, their connections and the position of the conserved GyrA-box motif. The deconvolution of two states of the DNA-binding/cleavage domain provides a better understanding of the allosteric movements of the enzyme complex. The local atomic resolution in the DNA-bound area reaching up to 3.0 Å enables the identification of the antibiotic density. Altogether, this study paves the way for the cryo-EM determination of gyrase complexes with antibiotics and opens perspectives for targeting conformational intermediates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4909.map.gz emd_4909.map.gz | 11.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4909-v30.xml emd-4909-v30.xml emd-4909.xml emd-4909.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4909_fsc.xml emd_4909_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_4909.png emd_4909.png | 59.2 KB | ||
| Masks |  emd_4909_msk_1.map emd_4909_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4909.cif.gz emd-4909.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4909 http://ftp.pdbj.org/pub/emdb/structures/EMD-4909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4909 | HTTPS FTP |
-Related structure data
| Related structure data |  6rksMC  4910C  4912C  4913C  4914C  4915C  6rkuC  6rkvC  6rkwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4909.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4909.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli DNA Gyrase DNA binding and cleavage domain. The TOPRIM insertion was masked away during refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4909_msk_1.map emd_4909_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1
| Entire | Name: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1 |
|---|---|
| Components |
|
-Supramolecule #1: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1
| Supramolecule | Name: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: The TOPRIM insertion was masked out during refinement. |
|---|
-Supramolecule #2: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1
| Supramolecule | Name: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: The TOPRIM insertion was masked out during refinement. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1
| Supramolecule | Name: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1 type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 Details: The TOPRIM insertion was masked out during refinement. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1
| Supramolecule | Name: DNA binding and cleavage domain of the E. coli DNA Gyrase in State 1 type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 Details: The TOPRIM insertion was masked out during refinement. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: DNA gyrase subunit A
| Macromolecule | Name: DNA gyrase subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA topoisomerase (ATP-hydrolysing) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 97.088586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDLAREITP VNIEEELKSS YLDYAMSVIV GRALPDVRDG LKPVHRRVLY AMNVLGNDWN KAYKKSARVV GDVIGKYHPH GDSAVYDTI VRMAQPFSLR YMLVDGQGNF GSIDGDSAAA MRYTEIRLAK IAHELMADLE KETVDFVDNY DGTEKIPDVM P TKIPNLLV ...String: MSDLAREITP VNIEEELKSS YLDYAMSVIV GRALPDVRDG LKPVHRRVLY AMNVLGNDWN KAYKKSARVV GDVIGKYHPH GDSAVYDTI VRMAQPFSLR YMLVDGQGNF GSIDGDSAAA MRYTEIRLAK IAHELMADLE KETVDFVDNY DGTEKIPDVM P TKIPNLLV NGSSGIAVGM ATNIPPHNLT EVINGCLAYI DDEDISIEGL MEHIPGPDFP TAAIINGRRG IEEAYRTGRG KV YIRARAE VEVDAKTGRE TIIVHEIPYQ VNKARLIEKI AELVKEKRVE GISALRDESD KDGMRIVIEV KRDAVGEVVL NNL YSQTQL QVSFGINMVA LHHGQPKIMN LKDIIAAFVR HRREVVTRRT IFELRKARDR AHILEALAVA LANIDPIIEL IRHA PTPAE AKTALVANPW QLGNVAAMLE RAGDDAARPE WLEPEFGVRD GLYYLTEQQA QAILDLRLQK LTGLEHEKLL DEYKE LLDQ IAELLRILGS ADRLMEVIRE ELELVREQFG DKRRTEITAN SADINLEDLI TQEDVVVTLS HQGYVKYQPL SEYEAQ RRG GKGKSAARIK EEDFIDRLLV ANTHDHILCF SSRGRVYSMK VYQLPEATRG ARGRPIVNLL PLEQDERITA ILPVTEF EE GVKVFMATAN GTVKKTVLTE FNRLRTAGKV AIKLVDGDEL IGVDLTSGED EVMLFSAEGK VVRFKESSVR AMGCNTTG V RGIRLGEGDK VVSLIVPRGD GAILTATQNG YGKRTAVAEY PTKSRATKGV ISIKVTERNG LVVGAVQVDD CDQIMMITD AGTLVRTRVS EISIVGRNTQ GVILIRTAED ENVVGLQRVA EPVDEEDLDT IDGSAAEGDD EIAPEVDVDD EPEEE UniProtKB: DNA gyrase subunit A |
-Macromolecule #2: DNA gyrase subunit B
| Macromolecule | Name: DNA gyrase subunit B / type: protein_or_peptide / ID: 2 / Details: GyrB subunit DNA Binding and cleavage domain / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA topoisomerase (ATP-hydrolysing) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90.073922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNSYDSSSI KVLKGLDAVR KRPGMYIGDT DDGTGLHHMV FEVVDNAIDE ALAGHCKEII VTIHADNSVS VQDDGRGIPT GIHPEEGVS AAEVIMTVLH AGGKFDDNSY KVSGGLHGVG VSVVNALSQK LELVIQREGK IHRQIYEHGV PQAPLAVTGE T EKTGTMVR ...String: MSNSYDSSSI KVLKGLDAVR KRPGMYIGDT DDGTGLHHMV FEVVDNAIDE ALAGHCKEII VTIHADNSVS VQDDGRGIPT GIHPEEGVS AAEVIMTVLH AGGKFDDNSY KVSGGLHGVG VSVVNALSQK LELVIQREGK IHRQIYEHGV PQAPLAVTGE T EKTGTMVR FWPSLETFTN VTEFEYEILA KRLRELSFLN SGVSIRLRDK RDGKEDHFHY EGGIKAFVEY LNKNKTPIHP NI FYFSTEK DGIGVEVALQ WNDGFQENIY CFTNNIPQRD GGTHLAGFRA AMTRTLNAYM DKEGYSKKAK VSATGDDARE GLI AVVSVK VPDPKFSSQT KDKLVSSEVK SAVEQQMNEL LAEYLLENPT DAKIVVGKII DAARAREAAR RAREMTRRKG ALDL AGLPG KLADCQERDP ALSELYLVEG DSAGGSAKQG RNRKNQAILP LKGKILNVEK ARFDKMLSSQ EVATLITALG CGIGR DEYN PDKLRYHSII IMTDADVDGS HIRTLLLTFF YRQMPEIVER GHVYIAQPPL YKVKKGKQEQ YIKDDEAMDQ YQISIA LDG ATLHTNASAP ALAGEALEKL VSEYNATQKM INRMERRYPK AMLKELIYQP TLTEADLSDE QTVTRWVNAL VSELNDK EQ HGSQWKFDVH TNAEQNLFEP IVRVRTHGVD TDYPLDHEFI TGGEYRRICT LGEKLRGLLE EDAFIERGER RQPVASFE Q ALDWLVKESR RGLSIQRYKG LGEMNPEQLW ETTMDPESRR MLRVTVKDAI AADQLFTTLM GDAVEPRRAF IEENALKAA NIDI UniProtKB: DNA gyrase subunit B |
-Macromolecule #3: DNA Strand 1
| Macromolecule | Name: DNA Strand 1 / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.288818 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DT)(DA)(DA)(DG)(DC)(DC) (DG)(DT)(DA)(DG) |
-Macromolecule #4: DNA Strand 2
| Macromolecule | Name: DNA Strand 2 / type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.506578 KDa |
| Sequence | String: (DG)(DT)(DA)(DC)(DC)(DT)(DA)(DC)(DG)(DG) (DC)(DT)(DT)(DA)(DT)(DG)(DA)(DT) |
-Macromolecule #5: (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethyla...
| Macromolecule | Name: (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione type: ligand / ID: 5 / Number of copies: 1 / Formula: JHN |
|---|---|
| Molecular weight | Theoretical: 448.518 Da |
| Chemical component information | 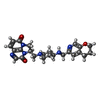 ChemComp-JHN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 3-40 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||||||
| Output model |  PDB-6rks: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)































