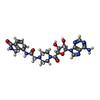
Entry Database : EMDB / ID : EMD-48313Title PARP1 ART in complex with HPF1 and EB47 PARP1 ART-HPF1-EB47, non-sharpened map Complex : Full-length human PARP1 bound to nicked DNA and in complex with HPF1 and Timeless fragmentProtein or peptide : Poly [ADP-ribose] polymerase 1Protein or peptide : Histone PARylation factor 1Ligand : 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 4.3 Å Sverzhinsky A / Pascal JM Funding support Organization Grant number Country Canadian Institutes of Health Research (CIHR) PJT374609 National Institutes of Health/National Cancer Institute (NIH/NCI) CA92584
Journal : To Be Published Title : Structure and dynamics of PARP1-HPF1 engaging nicked DNA suggests a mechanism for acute production of localized ADP-ribose modificationsAuthors : Sverzhinsky A / Xue H / Langelier M-F / Del Mundo J / Classen S / Hammel M / Rothenberg E / Pascal JM History Deposition Dec 14, 2024 - Header (metadata) release Jan 21, 2026 - Map release Jan 21, 2026 - Update Jan 21, 2026 - Current status Jan 21, 2026 Processing site : RCSB / Status : Released
Show all Show less
 Open data
Open data Basic information
Basic information
 Map data
Map data Sample
Sample Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Canada,
Canada,  United States, 2 items
United States, 2 items  Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Downloads & links
Downloads & links emd_48313.map.gz
emd_48313.map.gz EMDB map data format
EMDB map data format emd-48313-v30.xml
emd-48313-v30.xml emd-48313.xml
emd-48313.xml EMDB header
EMDB header emd_48313.png
emd_48313.png emd_48313_msk_1.map
emd_48313_msk_1.map Mask map
Mask map emd-48313.cif.gz
emd-48313.cif.gz emd_48313_additional_1.map.gz
emd_48313_additional_1.map.gz emd_48313_half_map_1.map.gz
emd_48313_half_map_1.map.gz emd_48313_half_map_2.map.gz
emd_48313_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-48313
http://ftp.pdbj.org/pub/emdb/structures/EMD-48313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48313
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48313 emd_48313_validation.pdf.gz
emd_48313_validation.pdf.gz EMDB validaton report
EMDB validaton report emd_48313_full_validation.pdf.gz
emd_48313_full_validation.pdf.gz emd_48313_validation.xml.gz
emd_48313_validation.xml.gz emd_48313_validation.cif.gz
emd_48313_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48313
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48313 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48313
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48313

 F&H Search
F&H Search Links
Links EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource Map
Map Download / File: emd_48313.map.gz / Format: CCP4 / Size: 25.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
Download / File: emd_48313.map.gz / Format: CCP4 / Size: 25.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_48313_msk_1.map
emd_48313_msk_1.map Sample components
Sample components Homo sapiens (human)
Homo sapiens (human) Homo sapiens (human)
Homo sapiens (human)
 Homo sapiens (human)
Homo sapiens (human)
 Processing
Processing Sample preparation
Sample preparation Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















































