[English] 日本語
 Yorodumi
Yorodumi- EMDB-48217: Structure of the human TWIK-2 potassium channel in complex with p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human TWIK-2 potassium channel in complex with pimozide | |||||||||
 Map data Map data | TWIK2 bound to pimozide | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Two-pore potassium channel K2P6 TWIK2 K2P channel pimozide drug NLRP3 inflammasome / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of lysosome size / Tandem of pore domain in a weak inwardly rectifying K+ channels (TWIK) / Phase 4 - resting membrane potential / potassium ion leak channel activity / regulation of resting membrane potential / inward rectifier potassium channel activity / negative regulation of systemic arterial blood pressure / outward rectifier potassium channel activity / positive regulation of NLRP3 inflammasome complex assembly / potassium channel activity ...regulation of lysosome size / Tandem of pore domain in a weak inwardly rectifying K+ channels (TWIK) / Phase 4 - resting membrane potential / potassium ion leak channel activity / regulation of resting membrane potential / inward rectifier potassium channel activity / negative regulation of systemic arterial blood pressure / outward rectifier potassium channel activity / positive regulation of NLRP3 inflammasome complex assembly / potassium channel activity / voltage-gated potassium channel complex / potassium ion transmembrane transport / potassium ion transport / late endosome membrane / lysosomal membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Khanra NK / Long SB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structure of the human TWIK-2 potassium channel and its inhibition by pimozide. Authors: Nandish K Khanra / Chongyuan Wang / Bryce D Delgado / Stephen B Long /  Abstract: The potassium channel TWIK-2 is crucial for ATP-induced activation of the NLRP3 inflammasome in macrophages. The channel is a member of the two-pore domain potassium (K2P) channel superfamily and an ...The potassium channel TWIK-2 is crucial for ATP-induced activation of the NLRP3 inflammasome in macrophages. The channel is a member of the two-pore domain potassium (K2P) channel superfamily and an emerging therapeutic target to mitigate severe inflammatory injury involving NLRP3 activation. We report the cryo-EM structure of human TWIK-2. In comparison to other K2P channels, the structure reveals an unusual "up" conformation of Tyr111 in the selectivity filter and a resulting SF1-P1 pocket behind the filter. Density for acyl chains is present in fenestrations within the transmembrane region that connects the central cavity of the pore to the lipid membrane. Despite its importance as a drug target, limited pharmacological tools are available for TWIK-2. A previous study suggested that the FDA-approved small molecule pimozide might inhibit TWIK-2. Using a reconstituted system, we show that pimozide directly inhibits the channel and we determine a cryo-EM structure of a complex with the drug. Pimozide displaces the acyl chains within the fenestrations and binds below the selectivity filter where it would impede ion permeation. The drug may access its binding site by lateral diffusion in the membrane, suggesting that other hydrophobic small molecules could have utility for inhibiting TWIK-2. The work defines the structure of TWIK-2 and provides a structural foundation for development of more specific inhibitors with potential utility as anti-inflammatory drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_48217.map.gz emd_48217.map.gz | 203.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-48217-v30.xml emd-48217-v30.xml emd-48217.xml emd-48217.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_48217_fsc.xml emd_48217_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_48217.png emd_48217.png | 67.1 KB | ||
| Filedesc metadata |  emd-48217.cif.gz emd-48217.cif.gz | 7 KB | ||
| Others |  emd_48217_half_map_1.map.gz emd_48217_half_map_1.map.gz emd_48217_half_map_2.map.gz emd_48217_half_map_2.map.gz | 200.5 MB 200.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-48217 http://ftp.pdbj.org/pub/emdb/structures/EMD-48217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48217 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-48217 | HTTPS FTP |
-Validation report
| Summary document |  emd_48217_validation.pdf.gz emd_48217_validation.pdf.gz | 833.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_48217_full_validation.pdf.gz emd_48217_full_validation.pdf.gz | 833.3 KB | Display | |
| Data in XML |  emd_48217_validation.xml.gz emd_48217_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_48217_validation.cif.gz emd_48217_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48217 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48217 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48217 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-48217 | HTTPS FTP |
-Related structure data
| Related structure data |  9melMC  9mekC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_48217.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_48217.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TWIK2 bound to pimozide | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.725 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: TWIK2 bound to pimozide
| File | emd_48217_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TWIK2 bound to pimozide | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: TWIK2 bound to pimozide
| File | emd_48217_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TWIK2 bound to pimozide | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human TWIK-2 potassium channel in complex with pimozide
| Entire | Name: human TWIK-2 potassium channel in complex with pimozide |
|---|---|
| Components |
|
-Supramolecule #1: human TWIK-2 potassium channel in complex with pimozide
| Supramolecule | Name: human TWIK-2 potassium channel in complex with pimozide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: human TWIK-2 potassium channel in complex with pimozide |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.74728 KDa |
-Macromolecule #1: Potassium channel subfamily K member 6
| Macromolecule | Name: Potassium channel subfamily K member 6 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.775113 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRRGALLAGA LAAYAAYLVL GALLVARLEG PHEARLRAEL ETLRAQLLQR SPCVAAPALD AFVERVLAAG RLGRVVLANA SGSANASDP AWDFASALFF ASTLITTVGY GYTTPLTDAG KAFSIAFALL GVPTTMLLLT ASAQRLSLLL THVPLSWLSM R WGWDPRRA ...String: MRRGALLAGA LAAYAAYLVL GALLVARLEG PHEARLRAEL ETLRAQLLQR SPCVAAPALD AFVERVLAAG RLGRVVLANA SGSANASDP AWDFASALFF ASTLITTVGY GYTTPLTDAG KAFSIAFALL GVPTTMLLLT ASAQRLSLLL THVPLSWLSM R WGWDPRRA ACWHLVALLG VVVTVCFLVP AVIFAHLEEA WSFLDAFYFC FISLSTIGLG DYVPGEAPGQ PYRALYKVLV TV YLFLGLV AMVLVLQTFR HVSDLHGLTE LILLPPPCPA SFNADEDDRV DILGPQPESH QQLSASSHTD YASIPR UniProtKB: Potassium channel subfamily K member 6 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimid...
| Macromolecule | Name: 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one type: ligand / ID: 3 / Number of copies: 1 / Formula: 1II |
|---|---|
| Molecular weight | Theoretical: 461.546 Da |
| Chemical component information | 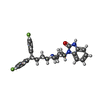 ChemComp-1II: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES, 150 mM KCl, pH 7.5, 0.01 % LMNG, 0.001 % CHS, 0.0033 % GDN, 0.5 mM pimozide | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 4.0 uL sample, blot time = 3.0 sec, blot force = 0. | |||||||||||||||||||||
| Details | Monodisperse recombinantly purified TWIK2 bound to pimozide |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 51.38 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 124.2 |
|---|---|
| Output model |  PDB-9mel: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































