+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4771 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae Niemann-Pick type C Related Protein 1 (NCR1) | ||||||||||||||||||
 Map data Map data | post-processed map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationIntestinal lipid absorption / LDL clearance / sterol binding / sterol transport / sphingolipid metabolic process / fungal-type vacuole membrane / transmembrane transporter activity / endoplasmic reticulum / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.2 Å | ||||||||||||||||||
 Authors Authors | Rawson S / Kidmose RT / Muench SP / Pedersen BP | ||||||||||||||||||
| Funding support |  Denmark, Denmark,  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins. Authors: Mikael B L Winkler / Rune T Kidmose / Maria Szomek / Katja Thaysen / Shaun Rawson / Stephen P Muench / Daniel Wüstner / Bjørn Panyella Pedersen /   Abstract: Niemann-Pick type C (NPC) proteins are essential for sterol homeostasis, believed to drive sterol integration into the lysosomal membrane before redistribution to other cellular membranes. Here, ...Niemann-Pick type C (NPC) proteins are essential for sterol homeostasis, believed to drive sterol integration into the lysosomal membrane before redistribution to other cellular membranes. Here, using a combination of crystallography, cryo-electron microscopy, and biochemical and in vivo studies on the Saccharomyces cerevisiae NPC system (NCR1 and NPC2), we present a framework for sterol membrane integration. Sterols are transferred between hydrophobic pockets of vacuolar NPC2 and membrane-protein NCR1. NCR1 has its N-terminal domain (NTD) positioned to deliver a sterol to a tunnel connecting NTD to the luminal membrane leaflet 50 Å away. A sterol is caught inside this tunnel during transport, and a proton-relay network of charged residues in the transmembrane region is linked to this tunnel supporting a proton-driven transport mechanism. We propose a model for sterol integration that clarifies the role of NPC proteins in this essential eukaryotic pathway and that rationalizes mutations in patients with Niemann-Pick disease type C. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4771.map.gz emd_4771.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4771-v30.xml emd-4771-v30.xml emd-4771.xml emd-4771.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4771_fsc.xml emd_4771_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4771.png emd_4771.png | 119.6 KB | ||
| Others |  emd_4771_additional.map.gz emd_4771_additional.map.gz emd_4771_additional_1.map.gz emd_4771_additional_1.map.gz emd_4771_half_map_1.map.gz emd_4771_half_map_1.map.gz emd_4771_half_map_2.map.gz emd_4771_half_map_2.map.gz | 26.5 MB 26.5 MB 31.4 MB 31.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4771 http://ftp.pdbj.org/pub/emdb/structures/EMD-4771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4771 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4771 | HTTPS FTP |
-Validation report
| Summary document |  emd_4771_validation.pdf.gz emd_4771_validation.pdf.gz | 381.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4771_full_validation.pdf.gz emd_4771_full_validation.pdf.gz | 380.7 KB | Display | |
| Data in XML |  emd_4771_validation.xml.gz emd_4771_validation.xml.gz | 13.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4771 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4771 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4771 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4771 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4771.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4771.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
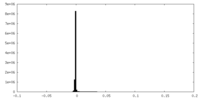
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: local resolution filtered.
| File | emd_4771_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered. | ||||||||||||
| Projections & Slices |
| ||||||||||||
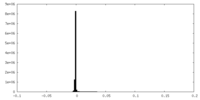
| Density Histograms |
-Additional map: local resolution filtered.
| File | emd_4771_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered. | ||||||||||||
| Projections & Slices |
| ||||||||||||
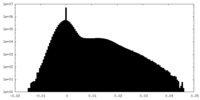
| Density Histograms |
-Half map: Half map 1
| File | emd_4771_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_4771_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
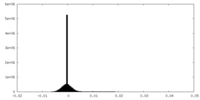
| Density Histograms |
- Sample components
Sample components
-Entire : NCR1
| Entire | Name: NCR1 |
|---|---|
| Components |
|
-Supramolecule #1: NCR1
| Supramolecule | Name: NCR1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #1: Niemann-Pick type C-related protein 1
| Macromolecule | Name: Niemann-Pick type C-related protein 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MNVLWIIALV GQLMRLVQGT ATCAMYGNCG KKSVFGNELP CPVPRSFEPP VLSDETSKLL VEVCGEEWKE VRYACCTKDQ VVALRDNLQK AQPLISSCPA CLKNFNNLFC HFTCAADQGR FVNITKVEKS KEDKDIVAEL DVFMNSSWAS EFYDSCKNIK FSATNGYAMD ...String: MNVLWIIALV GQLMRLVQGT ATCAMYGNCG KKSVFGNELP CPVPRSFEPP VLSDETSKLL VEVCGEEWKE VRYACCTKDQ VVALRDNLQK AQPLISSCPA CLKNFNNLFC HFTCAADQGR FVNITKVEKS KEDKDIVAEL DVFMNSSWAS EFYDSCKNIK FSATNGYAMD LIGGGAKNYS QFLKFLGDAK PMLGGSPFQI NYKYDLANEE KEWQEFNDEV YACDDAQYKC ACSDCQESCP HLKPLKDGVC KVGPLPCFSL SVLIFYTICA LFAFMWYYLC KRKKNGAMIV DDDIVPESGS LDESETNVFE SFNNETNFFN GKLANLFTKV GQFSVENPYK ILITTVFSIF VFSFIIFQYA TLETDPINLW VSKNSEKFKE KEYFDDNFGP FYRTEQIFVV NETGPVLSYE TLHWWFDVEN FITEELQSSE NIGYQDLCFR PTEDSTCVIE SFTQYFQGAL PNKDSWKREL QECGKFPVNC LPTFQQPLKT NLLFSDDDIL NAHAFVVTLL LTNHTQSANR WEERLEEYLL DLKVPEGLRI SFNTEISLEK ELNNNNDIST VAISYLMMFL YATWALRRKD GKTRLLLGIS GLLIVLASIV CAAGFLTLFG LKSTLIIAEV IPFLILAIGI DNIFLITHEY DRNCEQKPEY SIDQKIISAI GRMSPSILMS LLCQTGCFLI AAFVTMPAVH NFAIYSTVSV IFNGVLQLTA YVSILSLYEK RSNYKQITGN EETKESFLKT FYFKMLTQKR LIIIIFSAWF FTSLVFLPEI QFGLDQTLAV PQDSYLVDYF KDVYSFLNVG PPVYMVVKNL DLTKRQNQQK ICGKFTTCER DSLANVLEQE RHRSTITEPL ANWLDDYFMF LNPQNDQCCR LKKGTDEVCP PSFPSRRCET CFQQGSWNYN MSGFPEGKDF MEYLSIWINA PSDPCPLGGR APYSTALVYN ETSVSASVFR TAHHPLRSQK DFIQAYSDGV RISSSFPELD MFAYSPFYIF FVQYQTLGPL TLKLIGSAII LIFFISSVFL QNIRSSFLLA LVVTMIIVDI GALMALLGIS LNAVSLVNLI ICVGLGVEFC VHIVRSFTVV PSETKKDANS RVLYSLNTIG ESVIKGITLT KFIGVCVLAF AQSKIFDVFY FRMWFTLIIV AALHALLFLP ALLSLFGGES YRDDSIEAED LVPRGSGGGG SGGGGSGGHH HHHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)