[English] 日本語
 Yorodumi
Yorodumi- EMDB-47482: Torpedo muscle-type nicotinic acetylcholine receptor - Monoligand... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Torpedo muscle-type nicotinic acetylcholine receptor - Monoliganded State | |||||||||||||||||||||
 Map data Map data | Unsharpened map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Nicotinic acetylcholine receptor / Ion channel / Cys-loop receptor / Pentameric ligand-gated ion channel / Neurotransmitter-gated receptor / Ligand-gated ion channel / Primed state / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-gated monoatomic cation-selective channel activity / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / transmembrane signaling receptor activity / postsynaptic membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||||||||||||||
 Authors Authors | Thompson MJ / Nury H / Zarkadas E / Baenziger JE | |||||||||||||||||||||
| Funding support |  Canada, European Union, 6 items Canada, European Union, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2026 Journal: Science / Year: 2026Title: Asynchronous subunit transitions prime acetylcholine receptor activation. Authors: Mackenzie J Thompson / Christian J G Tessier / Anna Ananchenko / Camille Hénault / Johnathon R Emlaw / François Dehez / Eleftherios Zarkadas / Corrie J B daCosta / Hugues Nury / John E Baenziger /   Abstract: Communication at synapses is facilitated by postsynaptic receptors, which convert a chemical signal into an electrical response. For ligand-gated ion channels, agonist binding triggers rapid ...Communication at synapses is facilitated by postsynaptic receptors, which convert a chemical signal into an electrical response. For ligand-gated ion channels, agonist binding triggers rapid transitions through intermediate states leading to a transient open-pore conformation, with these transitions shaping the postsynaptic response. In this work, we determine structures of the muscle-type nicotinic acetylcholine receptor in unliganded, mono-liganded, and di-liganded states. Agonist binding to a single site stabilizes a closed structure where an entire principal agonist-binding subunit transitions to an active-like conformation, whereas the other unoccupied principal subunit remains inactive, albeit poised for activation. Uniting this intermediate structure with single-channel recordings informs a sequential activation mechanism where asynchronous subunit transitions prime the receptor for activation-a finding with implications for an entire superfamily of pentameric ligand-gated ion channels. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47482.map.gz emd_47482.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47482-v30.xml emd-47482-v30.xml emd-47482.xml emd-47482.xml | 34.5 KB 34.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47482_fsc.xml emd_47482_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_47482.png emd_47482.png | 30.5 KB | ||
| Masks |  emd_47482_msk_1.map emd_47482_msk_1.map emd_47482_msk_2.map emd_47482_msk_2.map | 64 MB 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-47482.cif.gz emd-47482.cif.gz | 9.3 KB | ||
| Others |  emd_47482_additional_1.map.gz emd_47482_additional_1.map.gz emd_47482_half_map_1.map.gz emd_47482_half_map_1.map.gz emd_47482_half_map_2.map.gz emd_47482_half_map_2.map.gz | 55.8 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47482 http://ftp.pdbj.org/pub/emdb/structures/EMD-47482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47482 | HTTPS FTP |
-Related structure data
| Related structure data |  9e3gMC  9e3eC  9e3fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47482.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47482.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.163 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_47482_msk_1.map emd_47482_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_47482_msk_2.map emd_47482_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: DeepEM enhancer map
| File | emd_47482_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEM enhancer map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A
| File | emd_47482_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_47482_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Torpedo muscle-type nicotinic acetylcholine receptor
| Entire | Name: Torpedo muscle-type nicotinic acetylcholine receptor |
|---|---|
| Components |
|
-Supramolecule #1: Torpedo muscle-type nicotinic acetylcholine receptor
| Supramolecule | Name: Torpedo muscle-type nicotinic acetylcholine receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 250 KDa |
-Macromolecule #1: Acetylcholine receptor subunit alpha
| Macromolecule | Name: Acetylcholine receptor subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.168164 KDa |
| Sequence | String: SEHETRLVAN LLENYNKVIR PVEHHTHFVD ITVGLQLIQL ISVDEVNQIV ETNVRLRQQW IDVRLRWNPA DYGGIKKIRL PSDDVWLPD LVLYNNADGD FAIVHMTKLL LDYTGKIMWT PPAIFKSYCE IIVTHFPFDQ QNCTMKLGIW TYDGTKVSIS P ESDRPDLS ...String: SEHETRLVAN LLENYNKVIR PVEHHTHFVD ITVGLQLIQL ISVDEVNQIV ETNVRLRQQW IDVRLRWNPA DYGGIKKIRL PSDDVWLPD LVLYNNADGD FAIVHMTKLL LDYTGKIMWT PPAIFKSYCE IIVTHFPFDQ QNCTMKLGIW TYDGTKVSIS P ESDRPDLS TFMESGEWVM KDYRGWKHWV YYTCCPDTPY LDITYHFIMQ RIPLYFVVNV IIPCLLFSFL TGLVFYLPTD SG EKMTLSI SVLLSLTVFL LVIVELIPST SSAVPLIGKY MLFTMIFVIS SIIITVVVIN THHRSPSTHT MPQWVRKIFI DTI PNVMFF STMKRASKEK QENKIFADDI DISDISGKQV TGEVIFQTPL IKNPDVKSAI EGVKYIAEHM KSDEESSNAA EEWK YVAMV IDHILLCVFM LICIIGTVSV FAGRLIELSQ EG UniProtKB: Acetylcholine receptor subunit alpha |
-Macromolecule #2: Acetylcholine receptor subunit beta
| Macromolecule | Name: Acetylcholine receptor subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.731773 KDa |
| Sequence | String: SVMEDTLLSV LFETYNPKVR PAQTVGDKVT VRVGLTLTNL LILNEKIEEM TTNVFLNLAW TDYRLQWDPA AYEGIKDLRI PSSDVWQPD IVLMNNNDGS FEITLHVNVL VQHTGAVSWQ PSAIYRSSCT IKVMYFPFDW QNCTMVFKSY TYDTSEVTLQ H ALDAKGER ...String: SVMEDTLLSV LFETYNPKVR PAQTVGDKVT VRVGLTLTNL LILNEKIEEM TTNVFLNLAW TDYRLQWDPA AYEGIKDLRI PSSDVWQPD IVLMNNNDGS FEITLHVNVL VQHTGAVSWQ PSAIYRSSCT IKVMYFPFDW QNCTMVFKSY TYDTSEVTLQ H ALDAKGER EVKEIVINKD AFTENGQWSI EHKPSRKNWR SDDPSYEDVT FYLIIQRKPL FYIVYTIIPC ILISILAILV FY LPPDAGE KMSLSISALL AVTVFLLLLA DKVPETSLSV PIIIRYLMFI MILVAFSVIL SVVVLNLHHR SPNTHTMPNW IRQ IFIETL PPFLWIQRPV TTPSPDSKPT IISRANDEYF IRKPAGDFVC PVDNARVAVQ PERLFSEMKW HLNGLTQPVT LPQD LKEAV EAIKYIAEQL ESASEFDDLK KDWQYVAMVA DRLFLYVFFV ICSIGTFSIF LDASHNVPPD NPFA UniProtKB: Acetylcholine receptor subunit beta |
-Macromolecule #3: Acetylcholine receptor subunit delta
| Macromolecule | Name: Acetylcholine receptor subunit delta / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.625711 KDa |
| Sequence | String: VNEEERLIND LLIVNKYNKH VRPVKHNNEV VNIALSLTLS NLISLKETDE TLTSNVWMDH AWYDHRLTWN ASEYSDISIL RLPPELVWI PDIVLQNNND GQYHVAYFCN VLVRPNGYVT WLPPAIFRSS CPINVLYFPF DWQNCSLKFT ALNYDANEIT M DLMTDTID ...String: VNEEERLIND LLIVNKYNKH VRPVKHNNEV VNIALSLTLS NLISLKETDE TLTSNVWMDH AWYDHRLTWN ASEYSDISIL RLPPELVWI PDIVLQNNND GQYHVAYFCN VLVRPNGYVT WLPPAIFRSS CPINVLYFPF DWQNCSLKFT ALNYDANEIT M DLMTDTID GKDYPIEWII IDPEAFTENG EWEIIHKPAK KNIYPDKFPN GTNYQDVTFY LIIRRKPLFY VINFITPCVL IS FLASLAF YLPAESGEKM STAISVLLAQ AVFLLLTSQR LPETALAVPL IGKYLMFIMS LVTGVIVNCG IVLNFHFRTP STH VLSTRV KQIFLEKLPR ILHMSRADES EQPDWQNDLK LRRSSSVGYI SKAQEYFNIK SRSELMFEKQ SERHGLVPRV TPRI GFGNN NENIAASDQL HDEIKSGIDS TNYIVKQIKE KNAYDEEVGN WNLVGQTIDR LSMFIITPVM VLGTIFIFVM GNFNH PPAK PFEGDPFDYS SDHPRCA UniProtKB: Acetylcholine receptor subunit delta |
-Macromolecule #4: Acetylcholine receptor subunit gamma
| Macromolecule | Name: Acetylcholine receptor subunit gamma / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.206574 KDa |
| Sequence | String: NEEGRLIEKL LGDYDKRIIP AKTLDHIIDV TLKLTLTNLI SLNEKEEALT TNVWIEIQWN DYRLSWNTSE YEGIDLVRIP SELLWLPDV VLENNVDGQF EVAYYANVLV YNDGSMYWLP PAIYRSTCPI AVTYFPFDWQ NCSLVFRSQT YNAHEVNLQL S AEEGEAVE ...String: NEEGRLIEKL LGDYDKRIIP AKTLDHIIDV TLKLTLTNLI SLNEKEEALT TNVWIEIQWN DYRLSWNTSE YEGIDLVRIP SELLWLPDV VLENNVDGQF EVAYYANVLV YNDGSMYWLP PAIYRSTCPI AVTYFPFDWQ NCSLVFRSQT YNAHEVNLQL S AEEGEAVE WIHIDPEDFT ENGEWTIRHR PAKKNYNWQL TKDDTDFQEI IFFLIIQRKP LFYIINIIAP CVLISSLVVL VY FLPAQAG GQKCTLSISV LLAQTIFLFL IAQKVPETSL NVPLIGKYLI FVMFVSMLIV MNCVIVLNVS LRTPNTHSLS EKI KHLFLG FLPKYLGMQL EPSEETPEKP QPRRRSSFGI MIKAEEYILK KPRSELMFEE QKDRHGLKRV NKMTSDIDIG TTVD LYKDL ANFAPEIKSC VEACNFIAKS TKEQNDSGSE NENWVLIGKV IDKACFWIAL LLFSIGTLAI FLTGHFNQVP EFPFP GDPR KYVP UniProtKB: Acetylcholine receptor subunit gamma |
-Macromolecule #10: ACETYLCHOLINE
| Macromolecule | Name: ACETYLCHOLINE / type: ligand / ID: 10 / Number of copies: 1 / Formula: ACH |
|---|---|
| Molecular weight | Theoretical: 146.207 Da |
| Chemical component information | 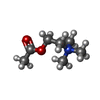 ChemComp-ACH: |
-Macromolecule #11: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 11 / Number of copies: 5 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.65 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: This map comes from particles from five datasets recorded in the presence of five sub-saturating concentrations of acetylcholine: 20, 50, 100, 250, and 500 nM. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | Sample was monodisperce but suffered from orientation bias. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






























































