[English] 日本語
 Yorodumi
Yorodumi- EMDB-4740: Atomic structure of potato virus X, the prototype of the Alphafle... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4740 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virus / Coat Protein / RNA | |||||||||
| Function / homology | Potexviruses and carlaviruses coat protein signature. / Potex/carlavirus coat protein / Viral coat protein / helical viral capsid / ribonucleoprotein complex / structural molecule activity / Coat protein / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Potato virus X Potato virus X | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Grinzato A / Kandiah E | |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2020 Journal: Nat Chem Biol / Year: 2020Title: Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family. Authors: Alessandro Grinzato / Eaazhisai Kandiah / Chiara Lico / Camilla Betti / Selene Baschieri / Giuseppe Zanotti /   Abstract: Potato virus X (PVX) is a positive-sense single-stranded RNA (ssRNA) filamentous plant virus belonging to the Alphaflexiviridae family, considered in recent years as a tool for nanotechnology ...Potato virus X (PVX) is a positive-sense single-stranded RNA (ssRNA) filamentous plant virus belonging to the Alphaflexiviridae family, considered in recent years as a tool for nanotechnology applications. We present the cryo-electron microscopy structure of the PVX particle at a resolution of 2.2 Å. The well-defined density of the coat proteins and of the genomic RNA allowed a detailed analysis of protein-RNA interactions, including those mediated by solvent molecules. The particle is formed by repeated segments made of 8.8 coat proteins, forming a left-handed helical structure. The RNA runs in an internal crevice along the virion, packaged in 5-nucleotide repeats in which the first four bases are stacked in the classical way, while the fifth is rotated and nearly perpendicular. The resolution of the structure described here suggests a mechanism for the virion assembly and potentially provides a platform for the rational design of antiviral compounds and for the use of PVX in nanotechnology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4740.map.gz emd_4740.map.gz | 166.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4740-v30.xml emd-4740-v30.xml emd-4740.xml emd-4740.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4740.png emd_4740.png | 58.9 KB | ||
| Filedesc metadata |  emd-4740.cif.gz emd-4740.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4740 http://ftp.pdbj.org/pub/emdb/structures/EMD-4740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4740 | HTTPS FTP |
-Related structure data
| Related structure data |  6r7gMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10705 (Title: structure of potato virus X, the prototype of the Alphaflexiviridae family EMPIAR-10705 (Title: structure of potato virus X, the prototype of the Alphaflexiviridae familyData size: 6.6 TB Data #1: Unaligned multi frame micographs of Potato Virus X dataset [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4740.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4740.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
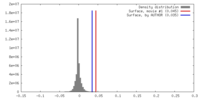
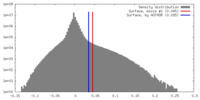
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Potato virus X
| Entire | Name:  Potato virus X Potato virus X |
|---|---|
| Components |
|
-Supramolecule #1: Potato virus X
| Supramolecule | Name: Potato virus X / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / NCBI-ID: 12183 / Sci species name: Potato virus X / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Potato virus X Potato virus X |
| Molecular weight | Theoretical: 22.519514 KDa |
| Sequence | String: ASGLFTIPDG DFFSTARAIV ASNAVATNED LSKIEAIWKD MKVPTDTMAQ AAWDLVRHCA DVGSSAQTEM IDTGPYSNGI SRARLAAAI KEVCTLRQFC MKYAPVVWNW MLTNNSPPAN WQAQGFKPEH KFAAFDFFNG VTNPAAIMPK EGLIRPPSEA E MNAAQTAA ...String: ASGLFTIPDG DFFSTARAIV ASNAVATNED LSKIEAIWKD MKVPTDTMAQ AAWDLVRHCA DVGSSAQTEM IDTGPYSNGI SRARLAAAI KEVCTLRQFC MKYAPVVWNW MLTNNSPPAN WQAQGFKPEH KFAAFDFFNG VTNPAAIMPK EGLIRPPSEA E MNAAQTAA FVKITKARAQ SNDFASLDAA VTRGRITGTT TAEAVVTLPP P UniProtKB: Coat protein |
-Macromolecule #2: polyU RNA
| Macromolecule | Name: polyU RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Potato virus X Potato virus X |
| Molecular weight | Theoretical: 21.080488 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUU |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 92 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 3.96 Å Applied symmetry - Helical parameters - Δ&Phi: 40.5 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 2.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 331130 |
|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Startup model | Type of model: INSILICO MODEL |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)