+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
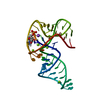
| Title | Structure of a Tick-Borne Flavivirus xrRNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Virus / RNA / xrRNA / Exonuclease | |||||||||
| Biological species |  Powassan virus Powassan virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Langeberg CJ / Kieft JS / Sherlock ME / Szucs MJ / Vicens Q | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Tick-borne flavivirus exoribonuclease-resistant RNAs contain a double loop structure. Authors: Conner J Langeberg / Matthew J Szucs / Madeline E Sherlock / Quentin Vicens / Jeffrey S Kieft /  Abstract: Viruses from the Flaviviridae family contain human relevant pathogens that generate subgenomic noncoding RNAs during infection using structured exoribonuclease resistant RNAs (xrRNAs). These xrRNAs ...Viruses from the Flaviviridae family contain human relevant pathogens that generate subgenomic noncoding RNAs during infection using structured exoribonuclease resistant RNAs (xrRNAs). These xrRNAs block progression of host cell's 5' to 3' exoribonucleases. The structures of several xrRNAs from mosquito-borne and insect-specific flaviviruses reveal a conserved fold in which a ring-like motif encircles the 5' end of the xrRNA. However, the xrRNAs found in tick-borne and no known vector flaviviruses have distinct characteristics, and their 3-D fold was unsolved. Here, we verify the presence of xrRNAs in the encephalitis-causing tick-borne Powassan Virus. We characterize their secondary structure and obtain a mid-resolution map of one of these xrRNAs using cryo-EM, revealing a unique double-loop ring element. Integrating these results with covariation analysis, biochemical data, and existing high-resolution structural information yields a model in which the core of the fold matches the previously solved xrRNA fold, but the expanded double loop ring is remodeled upon encountering the exoribonuclease. These results are representative of a broad class of xrRNAs and reveal a conserved strategy of structure-based exoribonuclease resistance achieved through a unique topology across a viral family of importance to global health. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47099.map.gz emd_47099.map.gz | 254 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47099-v30.xml emd-47099-v30.xml emd-47099.xml emd-47099.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_47099_fsc.xml emd_47099_fsc.xml | 23.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_47099.png emd_47099.png | 60.3 KB | ||
| Filedesc metadata |  emd-47099.cif.gz emd-47099.cif.gz | 5.5 KB | ||
| Others |  emd_47099_half_map_1.map.gz emd_47099_half_map_1.map.gz emd_47099_half_map_2.map.gz emd_47099_half_map_2.map.gz | 475.8 MB 475.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47099 http://ftp.pdbj.org/pub/emdb/structures/EMD-47099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47099 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_47099.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47099.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.788 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_47099_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_47099_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of a Tick-Borne Flavivirus xrRNA
| Entire | Name: Structure of a Tick-Borne Flavivirus xrRNA |
|---|---|
| Components |
|
-Supramolecule #1: Structure of a Tick-Borne Flavivirus xrRNA
| Supramolecule | Name: Structure of a Tick-Borne Flavivirus xrRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Powassan virus Powassan virus |
| Molecular weight | Theoretical: 142.44 KDa |
-Macromolecule #1: RNA (440-MER)
| Macromolecule | Name: RNA (440-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Powassan virus Powassan virus |
| Molecular weight | Theoretical: 142.224047 KDa |
| Sequence | String: UAGGGGGUGA UGUGGCAGCG CACUUGAUAU GGAUGCAGUU CACAGACUAA AUGUCGGUCG GGGAAGAUGU AUUCUUCUCA UAAGAUAUA GUCGGACCUC UCCUUAAUGG GAGCUAGCGG AUGAAGUGAU GCAACACUGG AGCCGCUGGG AACUAAUUUG U AUGCGAAA ...String: UAGGGGGUGA UGUGGCAGCG CACUUGAUAU GGAUGCAGUU CACAGACUAA AUGUCGGUCG GGGAAGAUGU AUUCUUCUCA UAAGAUAUA GUCGGACCUC UCCUUAAUGG GAGCUAGCGG AUGAAGUGAU GCAACACUGG AGCCGCUGGG AACUAAUUUG U AUGCGAAA GUAUAUUGAU UAGUUUUGGA GAAAGUUAUC AGGCAUGCAC CUGGUAGCUA GUCUUUAAAC CAAUAGAUUG CA UCGGUUU AAAAGGCAAG ACCGUCAAAU UGCGGGAAAG GGGUCAACAG CCGUUCAGUA CCAAGUCUCA GGGGAAACUU UGA GAUGGC CUUGCAAAGG GUAUGGUAAU AAGCUGACGG ACAUGGUCCU AACCACGCAG CCAAGUCCUA AGUCAAGUGA CGGG AAAAG GUCGUCCCCG ACGCAUCAUC CCUCUAUGCG AGACC |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 19 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)