[English] 日本語
 Yorodumi
Yorodumi- EMDB-4656: Localized reconstruction of archaeal virus SH1 spike calculated w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4656 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Localized reconstruction of archaeal virus SH1 spike calculated with C2 symmetry | |||||||||
 Map data Map data | Reconstruction of SH1 spike | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Haloarcula hispanica virus SH1 Haloarcula hispanica virus SH1 | |||||||||
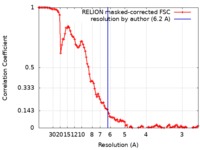
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | De Colibus L / Ilca SL / Stuart DIS / Huiskonen JT | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Assembly of complex viruses exemplified by a halophilic euryarchaeal virus. Authors: Luigi De Colibus / Elina Roine / Thomas S Walter / Serban L Ilca / Xiangxi Wang / Nan Wang / Alan M Roseman / Dennis Bamford / Juha T Huiskonen / David I Stuart /    Abstract: Many of the largest known viruses belong to the PRD1-adeno structural lineage characterised by conserved pseudo-hexameric capsomers composed of three copies of a single major capsid protein (MCP). ...Many of the largest known viruses belong to the PRD1-adeno structural lineage characterised by conserved pseudo-hexameric capsomers composed of three copies of a single major capsid protein (MCP). Here, by high-resolution cryo-EM analysis, we show that a class of archaeal viruses possess hetero-hexameric MCPs which mimic the PRD1-adeno lineage trimer. These hetero-hexamers are built from heterodimers and utilise a jigsaw-puzzle system of pegs and holes, and underlying minor capsid proteins, to assemble the capsid laterally from the 5-fold vertices. At these vertices proteins engage inwards with the internal membrane vesicle whilst 2-fold symmetric horn-like structures protrude outwards. The horns are assembled from repeated globular domains attached to a central spine, presumably facilitating multimeric attachment to the cell receptor. Such viruses may represent precursors of the main PRD1-adeno lineage, similarly engaging cell-receptors via 5-fold spikes and using minor proteins to define particle size. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4656.map.gz emd_4656.map.gz | 91.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4656-v30.xml emd-4656-v30.xml emd-4656.xml emd-4656.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4656_fsc.xml emd_4656_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_4656.png emd_4656.png | 84.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4656 http://ftp.pdbj.org/pub/emdb/structures/EMD-4656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4656 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4656.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4656.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of SH1 spike | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Haloarcula hispanica virus SH1
| Entire | Name:  Haloarcula hispanica virus SH1 Haloarcula hispanica virus SH1 |
|---|---|
| Components |
|
-Supramolecule #1: Haloarcula hispanica virus SH1
| Supramolecule | Name: Haloarcula hispanica virus SH1 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 326574 / Sci species name: Haloarcula hispanica virus SH1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 1000.0 Å / T number (triangulation number): 28 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)