[English] 日本語
 Yorodumi
Yorodumi- EMDB-45964: Cryo-EM structure of Tulane virus 9-6-17 variant capsid protein V... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Tulane virus 9-6-17 variant capsid protein VP1 9-14-18, DTT-treated | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tulane virus / capsid protein / VIRUS | |||||||||
| Function / homology | Calicivirus coat protein C-terminal / Calicivirus coat protein C-terminal / Calicivirus coat protein / Calicivirus coat protein / virion component / Viral coat protein subunit / host cell cytoplasm / Capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |  Tulane virus Tulane virus | |||||||||
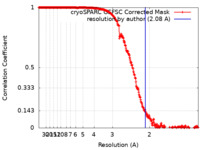
| Method | single particle reconstruction / cryo EM / Resolution: 2.08 Å | |||||||||
 Authors Authors | Sun C / Jiang W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2024 Journal: Biomolecules / Year: 2024Title: The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change. Authors: Chen Sun / Pengwei Huang / Xueyong Xu / Frank S Vago / Kunpeng Li / Thomas Klose / Xi Jason Jiang / Wen Jiang /  Abstract: Human noroviruses (HuNoVs) are a major cause of acute gastroenteritis, contributing significantly to annual foodborne illness cases. However, studying these viruses has been challenging due to ...Human noroviruses (HuNoVs) are a major cause of acute gastroenteritis, contributing significantly to annual foodborne illness cases. However, studying these viruses has been challenging due to limitations in tissue culture techniques for over four decades. Tulane virus (TV) has emerged as a crucial surrogate for HuNoVs due to its close resemblance in amino acid composition and the availability of a robust cell culture system. Initially isolated from rhesus macaques in 2008, TV represents a novel belonging to the genus. Its significance lies in sharing the same host cell receptor, histo-blood group antigen (HBGA), as HuNoVs. In this study, we introduce, through cryo-electron microscopy (cryo-EM), the structure of a specific TV variant (the 9-6-17 TV) that has notably lost its ability to bind to its receptor, B-type HBGA-a finding confirmed using an enzyme-linked immunosorbent assay (ELISA). These results offer a profound insight into the genetic modifications occurring in TV that are necessary for adaptation to cell culture environments. This research significantly contributes to advancing our understanding of the genetic changes that are pivotal to successful adaptation, shedding light on fundamental aspects of evolution. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45964.map.gz emd_45964.map.gz | 2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45964-v30.xml emd-45964-v30.xml emd-45964.xml emd-45964.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45964_fsc.xml emd_45964_fsc.xml | 33.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_45964.png emd_45964.png | 272.5 KB | ||
| Masks |  emd_45964_msk_1.map emd_45964_msk_1.map | 4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-45964.cif.gz emd-45964.cif.gz | 5.9 KB | ||
| Others |  emd_45964_half_map_1.map.gz emd_45964_half_map_1.map.gz emd_45964_half_map_2.map.gz emd_45964_half_map_2.map.gz | 3.7 GB 3.7 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45964 http://ftp.pdbj.org/pub/emdb/structures/EMD-45964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45964 | HTTPS FTP |
-Related structure data
| Related structure data |  9cvgMC  8vgrC  8vjrC  8vjsC  9cveC  9cvfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45964.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45964.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.692 Å | ||||||||||||||||||||||||||||||||||||
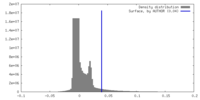
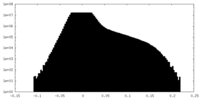
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45964_msk_1.map emd_45964_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
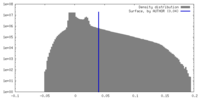
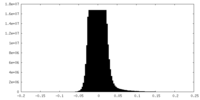
| Density Histograms |
-Half map: #2
| File | emd_45964_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
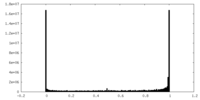
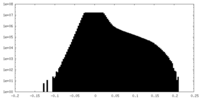
| Density Histograms |
-Half map: #1
| File | emd_45964_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
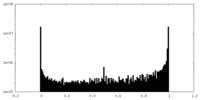
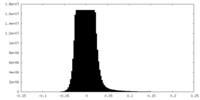
| Density Histograms |
- Sample components
Sample components
-Entire : Tulane virus
| Entire | Name:  Tulane virus Tulane virus |
|---|---|
| Components |
|
-Supramolecule #1: Tulane virus
| Supramolecule | Name: Tulane virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 512169 / Sci species name: Tulane virus / Virus type: VIROID / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tulane virus Tulane virus |
| Molecular weight | Theoretical: 57.933172 KDa |
| Sequence | String: MESSKTEQVT GATGITQSTV TAPLPEAVSS LSLAPTVNAL DPWVYLNQTE VPGGTFTVSS ATQPGSVLLE LEISPELNLY TSHLFRMYA GWSGGFSLKL LVAGNAFSAG KLIAAIIPPN IEVPNSAYLL TGFPHEILDF RTADSMEIIA PDIKNIDYHF R GDKLGKLV ...String: MESSKTEQVT GATGITQSTV TAPLPEAVSS LSLAPTVNAL DPWVYLNQTE VPGGTFTVSS ATQPGSVLLE LEISPELNLY TSHLFRMYA GWSGGFSLKL LVAGNAFSAG KLIAAIIPPN IEVPNSAYLL TGFPHEILDF RTADSMEIIA PDIKNIDYHF R GDKLGKLV VMVYSPLRST SADFEIEIKL TSAPLPDFKF TMLVPPIQNN ALPIWSIPQA PPYSMVNPRS PLTPVVELYI NS SYATCNH QLGRYTIYQG AIGNSTFNPS GAWTATCTAE AGSVTGHPNW RYALLDLPDN PTFDPTLPPV PRGFCDWGSG VKS GNKQHL VCFTGKKVEG GFQDVDTHMW DYGDNETVGL DNTYQRTIYI KDPSLEKDAQ YLVIPMGVSG AANDDTVQVA PNCY GSWDY APTVAPPLGE QFVWFRSQLP ASKTTTTSGV NSVPVNVNAL MSPDLMCSAY ASGFPLGKVA LLDYVLFGGS VVRQF KLYP EGYMTANTTG SNTGFIIPAD GYFRFNSWVS PSFMISSVVD LNLQTAVVFR UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 23.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)