[English] 日本語
 Yorodumi
Yorodumi- EMDB-4564: Cryo-EM reconstruction of heparin-induced 2N4R tau twister filaments -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4564 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of heparin-induced 2N4R tau twister filaments | |||||||||
 Map data Map data | None | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / tubulin complex / positive regulation of protein localization to synapse / phosphatidylinositol bisphosphate binding / generation of neurons / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / regulation of microtubule-based movement / regulation of chromosome organization / central nervous system neuron development / intracellular distribution of mitochondria / minor groove of adenine-thymine-rich DNA binding / lipoprotein particle binding / microtubule polymerization / negative regulation of mitochondrial membrane potential / regulation of microtubule polymerization / dynactin binding / apolipoprotein binding / main axon / protein polymerization / axolemma / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / negative regulation of mitochondrial fission / glial cell projection / neurofibrillary tangle assembly / positive regulation of axon extension / regulation of cellular response to heat / Activation of AMPK downstream of NMDARs / positive regulation of superoxide anion generation / positive regulation of protein localization / regulation of long-term synaptic depression / cellular response to brain-derived neurotrophic factor stimulus / positive regulation of microtubule polymerization / supramolecular fiber organization / synapse assembly / cytoplasmic microtubule organization / regulation of calcium-mediated signaling / somatodendritic compartment / axon cytoplasm / phosphatidylinositol binding / astrocyte activation / enzyme inhibitor activity / nuclear periphery / stress granule assembly / protein phosphatase 2A binding / regulation of microtubule cytoskeleton organization / cellular response to reactive oxygen species / Hsp90 protein binding / microglial cell activation / cellular response to nerve growth factor stimulus / synapse organization / PKR-mediated signaling / regulation of synaptic plasticity / protein homooligomerization / regulation of autophagy / response to lead ion / SH3 domain binding / microtubule cytoskeleton organization / memory / cytoplasmic ribonucleoprotein granule / neuron projection development / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / cellular response to heat / microtubule cytoskeleton / actin binding / growth cone / cell body / double-stranded DNA binding / protein-macromolecule adaptor activity / microtubule binding / sequence-specific DNA binding / dendritic spine / amyloid fibril formation / microtubule / learning or memory / neuron projection / membrane raft / negative regulation of gene expression / axon / neuronal cell body / DNA damage response / dendrite / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
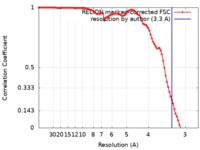
| Method | helical reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang W / Falcon B / Murzin AG / Fan J / Crowther RA / Goedert M / Scheres SHW | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. Authors: Wenjuan Zhang / Benjamin Falcon / Alexey G Murzin / Juan Fan / R Anthony Crowther / Michel Goedert / Sjors Hw Scheres /  Abstract: Assembly of microtubule-associated protein tau into filamentous inclusions underlies a range of neurodegenerative diseases. Tau filaments adopt different conformations in Alzheimer's and Pick's ...Assembly of microtubule-associated protein tau into filamentous inclusions underlies a range of neurodegenerative diseases. Tau filaments adopt different conformations in Alzheimer's and Pick's diseases. Here, we used cryo- and immuno- electron microscopy to characterise filaments that were assembled from recombinant full-length human tau with four (2N4R) or three (2N3R) microtubule-binding repeats in the presence of heparin. 2N4R tau assembles into multiple types of filaments, and the structures of three types reveal similar 'kinked hairpin' folds, in which the second and third repeats pack against each other. 2N3R tau filaments are structurally homogeneous, and adopt a dimeric core, where the third repeats of two tau molecules pack in a parallel manner. The heparin-induced tau filaments differ from those of Alzheimer's or Pick's disease, which have larger cores with different repeat compositions. Our results illustrate the structural versatility of amyloid filaments, and raise questions about the relevance of in vitro assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4564.map.gz emd_4564.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4564-v30.xml emd-4564-v30.xml emd-4564.xml emd-4564.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4564_fsc.xml emd_4564_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4564.png emd_4564.png | 24.9 KB | ||
| Masks |  emd_4564_msk_1.map emd_4564_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_4564_half_map_1.map.gz emd_4564_half_map_1.map.gz emd_4564_half_map_2.map.gz emd_4564_half_map_2.map.gz | 48.7 MB 48.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4564 http://ftp.pdbj.org/pub/emdb/structures/EMD-4564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4564 | HTTPS FTP |
-Related structure data
| Related structure data |  6qjmMC  4563C  4565C  4566C  6qjhC  6qjpC  6qjqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10243 (Title: Cryo-EM reconstruction of heparin-induced 2N4R tau filaments EMPIAR-10243 (Title: Cryo-EM reconstruction of heparin-induced 2N4R tau filamentsData size: 446.3 / Data #1: Aligned micrographs [micrographs - single frame] / Data #2: Raw movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4564.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4564.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4564_msk_1.map emd_4564_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map of helical reconstruction of heparin-induced 2N4R...
| File | emd_4564_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of helical reconstruction of heparin-induced 2N4R tau twister filaments | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The other half map of helical reconstruction of...
| File | emd_4564_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The other half map of helical reconstruction of heparin-induced 2N4R tau twister filaments | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : heparin-induced 2N4R tau twister filaments
| Entire | Name: heparin-induced 2N4R tau twister filaments |
|---|---|
| Components |
|
-Supramolecule #1: heparin-induced 2N4R tau twister filaments
| Supramolecule | Name: heparin-induced 2N4R tau twister filaments / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinant 2N4R tau protein was induced into filaments by adding heparin |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 45.85 kDa/nm |
-Macromolecule #1: Isoform Tau-F of Microtubule-associated protein tau
| Macromolecule | Name: Isoform Tau-F of Microtubule-associated protein tau / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKST PTAEDVTAPL VDEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA G HVTQARMV SKSKDGTGSD DKKAKGADGK TKIATPRGAA PPGQKGQANA ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKST PTAEDVTAPL VDEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA G HVTQARMV SKSKDGTGSD DKKAKGADGK TKIATPRGAA PPGQKGQANA TRIPAKTPPA PK TPPSSGE PPKSGDRSGY SSPGSPGTPG SRSRTPSLPT PPTREPKKVA VVRTPPKSPS SAK SRLQTA PVPMPDLKNV KSKIGSTENL KHQPGGGKVQ IINKKLDLSN VQSKCGSKDN IKHV PGGGS VQIVYKPVDL SKVTSKCGSL GNIHHKPGGG QVEVKSEKLD FKDRVQSKIG SLDNI THVP GGGNKKIETH KLTFRENAKA KTDHGAEIVY KSPVVSGDTS PRHLSNVSST GSIDMV DSP QLATLADEVS ASLAKQGL |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 20 mM Tris, pH 7.4, 100mM NaCl | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force: -12 ; Blot time: 4s. | |||||||||
| Details | Recombinant tau protein was induced into filaments by incubation with heparin at 37 degree celsius for 3 days |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Number real images: 717 / Average exposure time: 1.0 sec. / Average electron dose: 48.0 e/Å2 Details: Images were collected in movie-mode at 30 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.8 µm / Nominal defocus min: 1.7 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | A stack of three consecutive monomers was refined to preserve nearest-neighbour interactions for the middle chain. Side-chain clashes were detected using MOLPROBITY, and corrected by iterative cycles of real-space refinement in COOT and Fourier-space refinement in REFMAC and PHENIX. For each refined structure, separate model refinements were performed against a single half-map, and the resulting model was compared to the other half-map to confirm the absence of overfitting. |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 58.51 / Target criteria: Fourier shell correlation |
| Output model |  PDB-6qjm: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)