+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reconstituted P400 Subcomplex of the human TIP60 complex | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | complex / chromatin regulator / GENE REGULATION | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpiccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / sperm DNA condensation / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / histone chaperone activity / positive regulation of norepinephrine uptake / Regulation of CDH1 Function / establishment of protein localization to chromatin ...piccolo histone acetyltransferase complex / promoter-enhancer loop anchoring activity / telomerase RNA localization to Cajal body / sperm DNA condensation / regulation of DNA strand elongation / positive regulation of telomere maintenance in response to DNA damage / histone chaperone activity / positive regulation of norepinephrine uptake / Regulation of CDH1 Function / establishment of protein localization to chromatin / Formation of the polybromo-BAF (pBAF) complex / Formation of the non-canonical BAF (ncBAF) complex / Formation of the canonical BAF (cBAF) complex / Formation of the embryonic stem cell BAF (esBAF) complex / Formation of neuronal progenitor and neuronal BAF (npBAF and nBAF) / R2TP complex / dynein axonemal particle / bBAF complex / cellular response to cytochalasin B / neural retina development / npBAF complex / nBAF complex / brahma complex / regulation of transepithelial transport / RPAP3/R2TP/prefoldin-like complex / Swr1 complex / morphogenesis of a polarized epithelium / Formation of annular gap junctions / Formation of the dystrophin-glycoprotein complex (DGC) / structural constituent of postsynaptic actin cytoskeleton / GBAF complex / Gap junction degradation / Folding of actin by CCT/TriC / regulation of G0 to G1 transition / Cell-extracellular matrix interactions / protein localization to adherens junction / protein antigen binding / dense body / Tat protein binding / postsynaptic actin cytoskeleton / Ino80 complex / Prefoldin mediated transfer of substrate to CCT/TriC / RSC-type complex / blastocyst formation / regulation of double-strand break repair / regulation of nucleotide-excision repair / Adherens junctions interactions / RHOF GTPase cycle / chromatin-protein adaptor activity / adherens junction assembly / box C/D snoRNP assembly / apical protein localization / Sensory processing of sound by inner hair cells of the cochlea / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / tight junction / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of T cell differentiation / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / apical junction complex / positive regulation of double-strand break repair / spinal cord development / negative regulation of gene expression, epigenetic / maintenance of blood-brain barrier / regulation of norepinephrine uptake / regulation of chromosome organization / nitric-oxide synthase binding / transporter regulator activity / Transcriptional Regulation by E2F6 / cortical cytoskeleton / positive regulation of stem cell population maintenance / establishment or maintenance of cell polarity / NuA4 histone acetyltransferase complex / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / Recycling pathway of L1 / TFIID-class transcription factor complex binding / Regulation of MITF-M-dependent genes involved in pigmentation / regulation of DNA replication / MLL1 complex / brush border / regulation of G1/S transition of mitotic cell cycle / regulation of embryonic development / somatic stem cell population maintenance / EPH-ephrin mediated repulsion of cells / Telomere Extension By Telomerase / negative regulation of cell differentiation / spermatid development / protein folding chaperone complex / kinesin binding / enzyme-substrate adaptor activity / RHO GTPases Activate WASPs and WAVEs / regulation of synaptic vesicle endocytosis / positive regulation of myoblast differentiation / RNA polymerase II core promoter sequence-specific DNA binding / RHO GTPases activate IQGAPs / regulation of DNA repair / regulation of protein localization to plasma membrane / positive regulation of double-strand break repair via homologous recombination / EPHB-mediated forward signaling Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
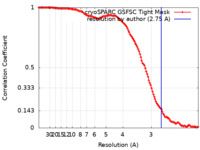
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | ||||||||||||||||||
 Authors Authors | Yang Z / Mameri A / Florez Ariza AJ / Cote J / Nogales E | ||||||||||||||||||
| Funding support |  United States, United States,  Canada, 5 items Canada, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex. Authors: Zhenlin Yang / Amel Mameri / Claudia Cattoglio / Catherine Lachance / Alfredo Jose Florez Ariza / Jie Luo / Jonathan Humbert / Deepthi Sudarshan / Arul Banerjea / Maxime Galloy / Amélie ...Authors: Zhenlin Yang / Amel Mameri / Claudia Cattoglio / Catherine Lachance / Alfredo Jose Florez Ariza / Jie Luo / Jonathan Humbert / Deepthi Sudarshan / Arul Banerjea / Maxime Galloy / Amélie Fradet-Turcotte / Jean-Philippe Lambert / Jeff A Ranish / Jacques Côté / Eva Nogales /   Abstract: The human nucleosome acetyltransferase of histone H4 (NuA4)/Tat-interactive protein, 60 kilodalton (TIP60) coactivator complex, a fusion of the yeast switch/sucrose nonfermentable related 1 (SWR1) ...The human nucleosome acetyltransferase of histone H4 (NuA4)/Tat-interactive protein, 60 kilodalton (TIP60) coactivator complex, a fusion of the yeast switch/sucrose nonfermentable related 1 (SWR1) and NuA4 complexes, both incorporates the histone variant H2A.Z into nucleosomes and acetylates histones H4, H2A, and H2A.Z to regulate gene expression and maintain genome stability. Our cryo-electron microscopy studies show that, within the NuA4/TIP60 complex, the E1A binding protein P400 (EP400) subunit serves as a scaffold holding the different functional modules in specific positions, creating a distinct arrangement of the actin-related protein (ARP) module. EP400 interacts with the transformation/transcription domain-associated protein (TRRAP) subunit by using a footprint that overlaps with that of the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex, preventing the formation of a hybrid complex. Loss of the TRRAP subunit leads to mislocalization of NuA4/TIP60, resulting in the redistribution of H2A.Z and its acetylation across the genome, emphasizing the dual functionality of NuA4/TIP60 as a single macromolecular assembly. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45206.map.gz emd_45206.map.gz | 167.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45206-v30.xml emd-45206-v30.xml emd-45206.xml emd-45206.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45206_fsc.xml emd_45206_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_45206.png emd_45206.png | 95.9 KB | ||
| Filedesc metadata |  emd-45206.cif.gz emd-45206.cif.gz | 10.5 KB | ||
| Others |  emd_45206_half_map_1.map.gz emd_45206_half_map_1.map.gz emd_45206_half_map_2.map.gz emd_45206_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45206 http://ftp.pdbj.org/pub/emdb/structures/EMD-45206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45206 | HTTPS FTP |
-Related structure data
| Related structure data |  9c57MC  9c47C  9c4bC  9c62C  9c6nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45206.map.gz / Format: CCP4 / Size: 204.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45206.map.gz / Format: CCP4 / Size: 204.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45206_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_45206_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Reconstituted P400 Subcomplex of the human TIP60 complex
+Supramolecule #1: Reconstituted P400 Subcomplex of the human TIP60 complex
+Macromolecule #1: RuvB-like 1
+Macromolecule #2: Enhancer of polycomb homolog 1
+Macromolecule #3: Actin-like protein 6A
+Macromolecule #4: RuvB-like 2
+Macromolecule #5: E1A-binding protein p400
+Macromolecule #6: DNA methyltransferase 1-associated protein 1
+Macromolecule #7: Actin, cytoplasmic 1
+Macromolecule #8: Vacuolar protein sorting-associated protein 72 homolog
+Macromolecule #9: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: 4D-STEM / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)