[English] 日本語
 Yorodumi
Yorodumi- EMDB-45177: Cryo-EM structure of the full-length human P2X4 receptor in the A... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the full-length human P2X4 receptor in the ATP-bound desensitized state | |||||||||
 Map data Map data | Locally sharpened map for the human P2X4 receptor in the ATP-bound desensitized state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane Protein / Ion Channel / Ligand-gated Ion Channel / P2X Receptor / Allosteric Antagonist | |||||||||
| Function / homology |  Function and homology information Function and homology informationsensory perception of touch / Platelet homeostasis / positive regulation of microglial cell migration / purinergic nucleotide receptor signaling pathway / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor activity / negative regulation of cardiac muscle hypertrophy / ligand-gated calcium channel activity / Elevation of cytosolic Ca2+ levels / positive regulation of prostaglandin secretion ...sensory perception of touch / Platelet homeostasis / positive regulation of microglial cell migration / purinergic nucleotide receptor signaling pathway / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor activity / negative regulation of cardiac muscle hypertrophy / ligand-gated calcium channel activity / Elevation of cytosolic Ca2+ levels / positive regulation of prostaglandin secretion / regulation of chemotaxis / tissue homeostasis / response to fluid shear stress / positive regulation of calcium ion transport / relaxation of cardiac muscle / positive regulation of endothelial cell chemotaxis / endothelial cell activation / response to ATP / cellular response to zinc ion / cellular response to ATP / behavioral response to pain / positive regulation of calcium ion transport into cytosol / positive regulation of blood vessel endothelial cell migration / membrane depolarization / regulation of cardiac muscle contraction / response to axon injury / Purinergic signaling in leishmaniasis infection / regulation of sodium ion transport / sensory perception of pain / positive regulation of calcium-mediated signaling / response to ischemia / apoptotic signaling pathway / calcium-mediated signaling / calcium ion transmembrane transport / regulation of blood pressure / positive regulation of nitric oxide biosynthetic process / cell junction / cell body / monoatomic ion transmembrane transport / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / postsynapse / cadherin binding / copper ion binding / signaling receptor binding / lysosomal membrane / perinuclear region of cytoplasm / signal transduction / extracellular exosome / zinc ion binding / ATP binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
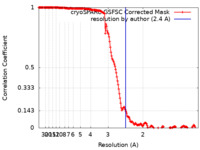
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Shi H / Ditter IA / Oken AC / Mansoor SE | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2025 Journal: Sci Adv / Year: 2025Title: Human P2X4 receptor gating is modulated by a stable cytoplasmic cap and a unique allosteric pocket. Authors: Haoyuan Shi / Ismayn A Ditter / Adam C Oken / Steven E Mansoor /  Abstract: P2X receptors (P2XRs) are adenosine 5'-triphosphate (ATP)-gated ion channels comprising homomeric and heteromeric trimers of seven subtypes (P2X1-P2X7) that confer different rates of desensitization. ...P2X receptors (P2XRs) are adenosine 5'-triphosphate (ATP)-gated ion channels comprising homomeric and heteromeric trimers of seven subtypes (P2X1-P2X7) that confer different rates of desensitization. The helical recoil model of P2XR desensitization proposes stability of the cytoplasmic cap sets the rate of desensitization, but timing of its formation is unclear for slow-desensitizing P2XRs. We report cryo-electron microscopy structures of full-length wild-type human P2X4 receptor in apo closed, antagonist-bound inhibited, and ATP-bound desensitized states. Because the apo closed and antagonist-bound inhibited state structures of this slow-desensitizing P2XR include an intact cytoplasmic cap while the ATP-bound desensitized state structure does not, the cytoplasmic cap is formed before agonist binding. Furthermore, structural and functional data suggest the cytoplasmic cap is stabilized by lipids to modulate desensitization, and P2X4 is modified by glycosylation and palmitoylation. Last, our antagonist-bound inhibited state structure reveals features specific to the allosteric ligand-binding pocket in human receptors that facilitates development of small-molecule modulators. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45177.map.gz emd_45177.map.gz | 228.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45177-v30.xml emd-45177-v30.xml emd-45177.xml emd-45177.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45177_fsc.xml emd_45177_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_45177.png emd_45177.png | 110.6 KB | ||
| Masks |  emd_45177_msk_1.map emd_45177_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45177.cif.gz emd-45177.cif.gz | 6.7 KB | ||
| Others |  emd_45177_additional_1.map.gz emd_45177_additional_1.map.gz emd_45177_additional_2.map.gz emd_45177_additional_2.map.gz emd_45177_half_map_1.map.gz emd_45177_half_map_1.map.gz emd_45177_half_map_2.map.gz emd_45177_half_map_2.map.gz | 229.7 MB 447.8 MB 440.3 MB 440.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45177 http://ftp.pdbj.org/pub/emdb/structures/EMD-45177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45177 | HTTPS FTP |
-Related structure data
| Related structure data |  9c48MC  9bqhC  9bqiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45177.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45177.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally sharpened map for the human P2X4 receptor in the ATP-bound desensitized state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.648 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45177_msk_1.map emd_45177_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened map for the human P2X4 receptor in...
| File | emd_45177_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map for the human P2X4 receptor in the ATP-bound desensitized state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map for the human P2X4 receptor in...
| File | emd_45177_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map for the human P2X4 receptor in the ATP-bound desensitized state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A for the human P2X4 receptor...
| File | emd_45177_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for the human P2X4 receptor in the ATP-bound desensitized state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B for the human P2X4 receptor...
| File | emd_45177_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for the human P2X4 receptor in the ATP-bound desensitized state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Membrane protein
| Entire | Name: Membrane protein |
|---|---|
| Components |
|
-Supramolecule #1: Membrane protein
| Supramolecule | Name: Membrane protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: P2X purinoceptor 4
| Macromolecule | Name: P2X purinoceptor 4 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.415 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGCCAALAA FLFEYDTPRI VLIRSRKVGL MNRAVQLLIL AYVIGWVFVW EKGYQETDSV VSSVTTKVKG VAVTNTSKLG FRIWDVADY VIPAQEENSL FVMTNVILTM NQTQGLCPEI PDATTVCKSD ASCTAGSAGT HSNGVSTGRC VAFNGSVKTC E VAAWCPVE ...String: MAGCCAALAA FLFEYDTPRI VLIRSRKVGL MNRAVQLLIL AYVIGWVFVW EKGYQETDSV VSSVTTKVKG VAVTNTSKLG FRIWDVADY VIPAQEENSL FVMTNVILTM NQTQGLCPEI PDATTVCKSD ASCTAGSAGT HSNGVSTGRC VAFNGSVKTC E VAAWCPVE DDTHVPQPAF LKAAENFTLL VKNNIWYPKF NFSKRNILPN ITTTYLKSCI YDAKTDPFCP IFRLGKIVEN AG HSFQDMA VEGGIMGIQV NWDCNLDRAA SLCLPRYSFR RLDTRDVEHN VSPGYNFRFA KYYRDLAGNE QRTLIKAYGI RFD IIVFGK AGKFDIIPTM INIGSGLALL GMATVLCDII VLYCMKKRLY YREKKYKYVE DYEQGLASEL DQ UniProtKB: P2X purinoceptor 4 |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 249 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 15582 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)