+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of a Tm1C Fibril | |||||||||
 Map data Map data | EM density map of the class A structure from Relion post-processing, using a 10% central Z length mask. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN FIBRIL | |||||||||
| Function / homology | Tropomyosin / Tropomyosin / GH09289p Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
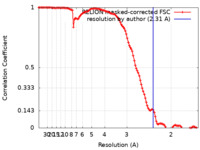
| Method | helical reconstruction / cryo EM / Resolution: 2.31 Å | |||||||||
 Authors Authors | Fonda BD / Kato M / Li Y / Murray DT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2024 Journal: Protein Sci / Year: 2024Title: Cryo-EM and solid state NMR together provide a more comprehensive structural investigation of protein fibrils. Authors: Blake D Fonda / Masato Kato / Yang Li / Dylan T Murray /  Abstract: The tropomyosin 1 isoform I/C C-terminal domain (Tm1-LC) fibril structure is studied jointly with cryogenic electron microscopy (cryo-EM) and solid state nuclear magnetic resonance (NMR). This study ...The tropomyosin 1 isoform I/C C-terminal domain (Tm1-LC) fibril structure is studied jointly with cryogenic electron microscopy (cryo-EM) and solid state nuclear magnetic resonance (NMR). This study demonstrates the complementary nature of these two structural biology techniques. Chemical shift assignments from solid state NMR are used to determine the secondary structure at the level of individual amino acids, which is faithfully seen in cryo-EM reconstructions. Additionally, solid state NMR demonstrates that the region not observed in the reconstructed cryo-EM density is primarily in a highly mobile random coil conformation rather than adopting multiple rigid conformations. Overall, this study illustrates the benefit of investigations combining cryo-EM and solid state NMR to investigate protein fibril structure. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
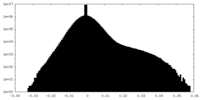
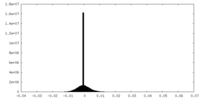
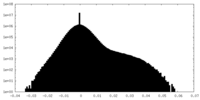
| Map data |  emd_45130.map.gz emd_45130.map.gz | 5.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45130-v30.xml emd-45130-v30.xml emd-45130.xml emd-45130.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45130_fsc.xml emd_45130_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_45130.png emd_45130.png | 144.7 KB | ||
| Masks |  emd_45130_msk_1.map emd_45130_msk_1.map emd_45130_msk_2.map emd_45130_msk_2.map | 125 MB 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-45130.cif.gz emd-45130.cif.gz | 6 KB | ||
| Others |  emd_45130_additional_1.map.gz emd_45130_additional_1.map.gz emd_45130_half_map_1.map.gz emd_45130_half_map_1.map.gz emd_45130_half_map_2.map.gz emd_45130_half_map_2.map.gz | 6.7 MB 98.5 MB 98.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45130 http://ftp.pdbj.org/pub/emdb/structures/EMD-45130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45130 | HTTPS FTP |
-Validation report
| Summary document |  emd_45130_validation.pdf.gz emd_45130_validation.pdf.gz | 841.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_45130_full_validation.pdf.gz emd_45130_full_validation.pdf.gz | 840.7 KB | Display | |
| Data in XML |  emd_45130_validation.xml.gz emd_45130_validation.xml.gz | 18.7 KB | Display | |
| Data in CIF |  emd_45130_validation.cif.gz emd_45130_validation.cif.gz | 24.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45130 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45130 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45130 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45130 | HTTPS FTP |
-Related structure data
| Related structure data |  9c1uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_45130.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45130.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM density map of the class A structure from Relion post-processing, using a 10% central Z length mask. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
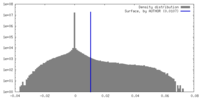
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_45130_msk_1.map emd_45130_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_45130_msk_2.map emd_45130_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: EM density map of the class A structure...
| File | emd_45130_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM density map of the class A structure from Relion post-processing, using a 20% central Z length mask. This map is used for model building and refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The first unfiltered half map used in Relion post-processing.
| File | emd_45130_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first unfiltered half map used in Relion post-processing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second unfiltered half map used in Relion post-processing.
| File | emd_45130_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second unfiltered half map used in Relion post-processing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tropomyosin 1 I/C C-terminal Domain
| Entire | Name: Tropomyosin 1 I/C C-terminal Domain |
|---|---|
| Components |
|
-Supramolecule #1: Tropomyosin 1 I/C C-terminal Domain
| Supramolecule | Name: Tropomyosin 1 I/C C-terminal Domain / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.41557 kDa/nm |
-Macromolecule #1: Tropomyosin 1 I/C
| Macromolecule | Name: Tropomyosin 1 I/C / type: protein_or_peptide / ID: 1 / Details: SY-tagged Tm1 I/C alternative isoform / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.420535 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SYSSIKLSNN NNNSNSNNIE ISKSESCNAS DIGGTNNNNA SRTIASAAVG EETSTLSSTS HEHNNNPNND T UniProtKB: GH09289p |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)